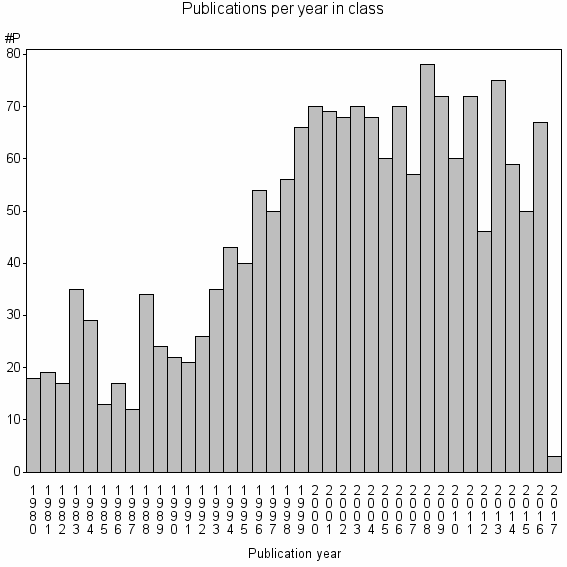
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 4671 | 1745 | 52.6 | 83% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 160 | 2 | WERNER SYNDROME//DNA POLYMERASE//DNA REPAIR | 22798 |
| 4671 | 1 | RAD51//RAD52//HOMOLOGOUS RECOMBINATION | 1745 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RAD51 | authKW | 849061 | 9% | 32% | 152 |
| 2 | RAD52 | authKW | 332051 | 3% | 43% | 44 |
| 3 | HOMOLOGOUS RECOMBINATION | authKW | 326193 | 12% | 9% | 208 |
| 4 | RAD51 PARALOGS | authKW | 187460 | 1% | 71% | 15 |
| 5 | RAD54 | authKW | 134370 | 1% | 40% | 19 |
| 6 | SRS2 | authKW | 123196 | 1% | 54% | 13 |
| 7 | HUMAN RAD51 | authKW | 107168 | 0% | 88% | 7 |
| 8 | SHU COMPLEX | authKW | 104982 | 0% | 100% | 6 |
| 9 | DOUBLE STRAND BREAK REPAIR | authKW | 88642 | 2% | 13% | 39 |
| 10 | ROSENSTIEL | address | 75831 | 1% | 19% | 23 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 16451 | 40% | 0% | 706 |
| 2 | Biochemistry & Molecular Biology | 6450 | 51% | 0% | 884 |
| 3 | Cell Biology | 5649 | 30% | 0% | 522 |
| 4 | Toxicology | 1261 | 9% | 0% | 152 |
| 5 | Oncology | 236 | 8% | 0% | 142 |
| 6 | Biology | 177 | 4% | 0% | 62 |
| 7 | Biotechnology & Applied Microbiology | 165 | 6% | 0% | 98 |
| 8 | Mycology | 93 | 1% | 0% | 19 |
| 9 | Developmental Biology | 65 | 2% | 0% | 28 |
| 10 | Biophysics | 30 | 3% | 0% | 47 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ROSENSTIEL | 75831 | 1% | 19% | 23 |
| 2 | SIGRR | 34994 | 0% | 100% | 2 |
| 3 | STRUCT BIOL SHINJUKU KU | 34994 | 0% | 100% | 2 |
| 4 | UMR 217 | 27974 | 1% | 13% | 12 |
| 5 | ANAL STRUCT | 26243 | 0% | 50% | 3 |
| 6 | ROSENTIEL MS029 | 23328 | 0% | 67% | 2 |
| 7 | RADIAT GENOME STABIL UNIT | 21305 | 1% | 7% | 17 |
| 8 | BIOL MICROBIAL GENET | 20402 | 0% | 17% | 7 |
| 9 | SECT MOL CELLULAR BIOL | 19856 | 2% | 4% | 28 |
| 10 | MOL RADIOLBUNKYO KU | 19680 | 0% | 38% | 3 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 45769 | 4% | 4% | 73 |
| 2 | GENETICS | 40962 | 10% | 1% | 173 |
| 3 | CURRENT GENETICS | 21924 | 4% | 2% | 68 |
| 4 | MOLECULAR AND CELLULAR BIOLOGY | 12976 | 7% | 1% | 129 |
| 5 | MUTATION RESEARCH-DNA REPAIR | 9167 | 1% | 3% | 16 |
| 6 | MOLECULAR CELL | 5596 | 2% | 1% | 42 |
| 7 | NUCLEIC ACIDS RESEARCH | 5520 | 6% | 0% | 113 |
| 8 | PLOS GENETICS | 4137 | 2% | 1% | 38 |
| 9 | MUTATION RESEARCH-FUNDAMENTAL AND MOLECULAR MECHANISMS OF MUTAGENESIS | 3729 | 2% | 1% | 29 |
| 10 | MOLECULAR AND GENERAL GENETICS | 3017 | 2% | 1% | 29 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RAD51 | 849061 | 9% | 32% | 152 | Search RAD51 | Search RAD51 |
| 2 | RAD52 | 332051 | 3% | 43% | 44 | Search RAD52 | Search RAD52 |
| 3 | HOMOLOGOUS RECOMBINATION | 326193 | 12% | 9% | 208 | Search HOMOLOGOUS+RECOMBINATION | Search HOMOLOGOUS+RECOMBINATION |
| 4 | RAD51 PARALOGS | 187460 | 1% | 71% | 15 | Search RAD51+PARALOGS | Search RAD51+PARALOGS |
| 5 | RAD54 | 134370 | 1% | 40% | 19 | Search RAD54 | Search RAD54 |
| 6 | SRS2 | 123196 | 1% | 54% | 13 | Search SRS2 | Search SRS2 |
| 7 | HUMAN RAD51 | 107168 | 0% | 88% | 7 | Search HUMAN+RAD51 | Search HUMAN+RAD51 |
| 8 | SHU COMPLEX | 104982 | 0% | 100% | 6 | Search SHU+COMPLEX | Search SHU+COMPLEX |
| 9 | DOUBLE STRAND BREAK REPAIR | 88642 | 2% | 13% | 39 | Search DOUBLE+STRAND+BREAK+REPAIR | Search DOUBLE+STRAND+BREAK+REPAIR |
| 10 | SINGLE STRAND ANNEALING | 72947 | 1% | 30% | 14 | Search SINGLE+STRAND+ANNEALING | Search SINGLE+STRAND+ANNEALING |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SYMINGTON, LS , (2002) ROLE OF RAD52 EPISTASIS GROUP GENES IN HOMOLOGOUS RECOMBINATION AND DOUBLE-STRAND BREAK REPAIR.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 66. ISSUE 4. P. 630 -+ | 210 | 51% | 633 |
| 2 | SYMINGTON, LS , ROTHSTEIN, R , LISBY, M , (2014) MECHANISMS AND REGULATION OF MITOTIC RECOMBINATION IN SACCHAROMYCES CEREVISIAE.GENETICS. VOL. 198. ISSUE 3. P. 795 -835 | 200 | 40% | 34 |
| 3 | DUDAS, A , CHOVANEC, M , (2004) DNA DOUBLE-STRAND BREAK REPAIR BY HOMOLOGOUS RECOMBINATION.MUTATION RESEARCH-REVIEWS IN MUTATION RESEARCH. VOL. 566. ISSUE 2. P. 131-167 | 196 | 52% | 102 |
| 4 | ZHAO, XL , SPIREK, M , ALTMANNOVA, V , KREJCI, L , (2012) HOMOLOGOUS RECOMBINATION AND ITS REGULATION.NUCLEIC ACIDS RESEARCH. VOL. 40. ISSUE 13. P. 5795 -5818 | 131 | 41% | 165 |
| 5 | PAQUES, F , HABER, JE , (1999) MULTIPLE PATHWAYS OF RECOMBINATION INDUCED BY DOUBLE-STRAND BREAKS IN SACCHAROMYCES CEREVISIAE.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 63. ISSUE 2. P. 349-+ | 152 | 30% | 1478 |
| 6 | HANAMSHET, K , MAZINA, OM , MAZIN, AV , (2016) REAPPEARANCE FROM OBSCURITY: MAMMALIAN RAD52 IN HOMOLOGOUS RECOMBINATION.GENES. VOL. 7. ISSUE 9. P. - | 83 | 71% | 0 |
| 7 | CEBALLOS, SJ , HEYER, WD , (2011) FUNCTIONS OF THE SNF2/SWI2 FAMILY RAD54 MOTOR PROTEIN IN HOMOLOGOUS RECOMBINATION.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1809. ISSUE 9. P. 509 -523 | 93 | 58% | 46 |
| 8 | VAN DEN BOSCH, M , LOHMAN, PHM , PASTINK, A , (2002) DNA DOUBLE-STRAND BREAK REPAIR BY HOMOLOGOUS RECOMBINATION.BIOLOGICAL CHEMISTRY. VOL. 383. ISSUE 6. P. 873-892 | 114 | 59% | 99 |
| 9 | PRADO, FL , CORTES-LEDESMA, F , HUERTAS, P , AGUILERA, A , (2003) MITOTIC RECOMBINATION IN SACCHAROMYCES CEREVISIAE.CURRENT GENETICS. VOL. 42. ISSUE 4. P. 185-198 | 97 | 67% | 57 |
| 10 | FILIPPO, JS , SUNG, P , KLEIN, H , (2008) MECHANISM OF EUKARYOTIC HOMOLOGOUS RECOMBINATION.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 77. ISSUE . P. 229 -257 | 98 | 48% | 126 |
Classes with closest relation at Level 1 |