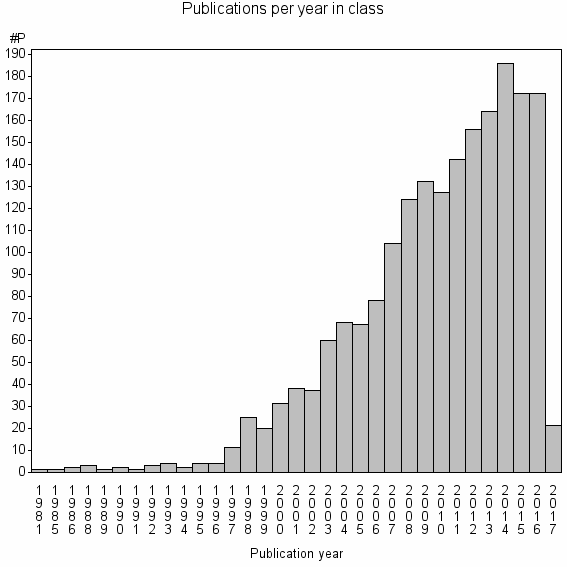
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 3535 | 1963 | 54.5 | 95% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 271 | 2 | INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH//ATAXIA TELANGIECTASIA | 19288 |
| 3535 | 1 | NIJMEGEN BREAKAGE SYNDROME//NBS1//MRE11 | 1963 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | NIJMEGEN BREAKAGE SYNDROME | authKW | 841204 | 4% | 72% | 75 |
| 2 | NBS1 | authKW | 812876 | 4% | 62% | 84 |
| 3 | MRE11 | authKW | 493798 | 3% | 50% | 63 |
| 4 | 53BP1 | authKW | 474213 | 4% | 42% | 72 |
| 5 | RNF168 | authKW | 244119 | 1% | 83% | 19 |
| 6 | MDC1 | authKW | 237079 | 1% | 61% | 25 |
| 7 | MRN COMPLEX | authKW | 218491 | 1% | 59% | 24 |
| 8 | RNF8 | authKW | 210953 | 1% | 59% | 23 |
| 9 | RAD50 | authKW | 205824 | 2% | 44% | 30 |
| 10 | RAP80 | authKW | 179966 | 1% | 64% | 18 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Cell Biology | 16727 | 47% | 0% | 924 |
| 2 | Biochemistry & Molecular Biology | 6163 | 47% | 0% | 925 |
| 3 | Genetics & Heredity | 4927 | 22% | 0% | 425 |
| 4 | Oncology | 1072 | 14% | 0% | 273 |
| 5 | Toxicology | 660 | 6% | 0% | 122 |
| 6 | Biophysics | 335 | 6% | 0% | 114 |
| 7 | Developmental Biology | 209 | 2% | 0% | 48 |
| 8 | Biology | 123 | 3% | 0% | 58 |
| 9 | Radiology, Nuclear Medicine & Medical Imaging | 22 | 3% | 0% | 52 |
| 10 | Medicine, Research & Experimental | 4 | 2% | 0% | 36 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENOME REPAIR DYNAM | 102077 | 1% | 41% | 16 |
| 2 | RADIAT BIOL DNA REPAIR | 74100 | 0% | 53% | 9 |
| 3 | GENOTOX ST S | 72777 | 1% | 20% | 23 |
| 4 | GENOME DAMAGE STABIL | 57253 | 2% | 8% | 46 |
| 5 | GENOME STABIL | 57023 | 1% | 15% | 24 |
| 6 | DAVID INEZ MYERS CANC GENET | 35344 | 0% | 45% | 5 |
| 7 | STATE NAT BIOMIMET DRUGSBEIJING KEY LA | 34994 | 0% | 75% | 3 |
| 8 | GURDON | 32346 | 1% | 8% | 27 |
| 9 | BIOCHEM MOL BIOL CARCINOGENESIS | 31107 | 0% | 100% | 2 |
| 10 | GASTROENTEROL HEPATOL CLIN IMMUNOL | 31107 | 0% | 100% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 48859 | 4% | 4% | 80 |
| 2 | MOLECULAR CELL | 16380 | 4% | 1% | 76 |
| 3 | COLD SPRING HARBOR SYMPOSIA ON QUANTITATIVE BIOLOGY SERIES | 13248 | 3% | 1% | 65 |
| 4 | NATURE STRUCTURAL & MOLECULAR BIOLOGY | 8760 | 2% | 2% | 35 |
| 5 | NATURE CELL BIOLOGY | 6241 | 2% | 1% | 33 |
| 6 | GENES & DEVELOPMENT | 3799 | 2% | 1% | 43 |
| 7 | EMBO REPORTS | 3766 | 1% | 1% | 23 |
| 8 | PLOS GENETICS | 3107 | 2% | 1% | 35 |
| 9 | CELL REPORTS | 2938 | 1% | 1% | 25 |
| 10 | NUCLEIC ACIDS RESEARCH | 2855 | 4% | 0% | 87 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | NIJMEGEN BREAKAGE SYNDROME | 841204 | 4% | 72% | 75 | Search NIJMEGEN+BREAKAGE+SYNDROME | Search NIJMEGEN+BREAKAGE+SYNDROME |
| 2 | NBS1 | 812876 | 4% | 62% | 84 | Search NBS1 | Search NBS1 |
| 3 | MRE11 | 493798 | 3% | 50% | 63 | Search MRE11 | Search MRE11 |
| 4 | 53BP1 | 474213 | 4% | 42% | 72 | Search 53BP1 | Search 53BP1 |
| 5 | RNF168 | 244119 | 1% | 83% | 19 | Search RNF168 | Search RNF168 |
| 6 | MDC1 | 237079 | 1% | 61% | 25 | Search MDC1 | Search MDC1 |
| 7 | MRN COMPLEX | 218491 | 1% | 59% | 24 | Search MRN+COMPLEX | Search MRN+COMPLEX |
| 8 | RNF8 | 210953 | 1% | 59% | 23 | Search RNF8 | Search RNF8 |
| 9 | RAD50 | 205824 | 2% | 44% | 30 | Search RAD50 | Search RAD50 |
| 10 | RAP80 | 179966 | 1% | 64% | 18 | Search RAP80 | Search RAP80 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SYMINGTON, LS , (2016) MECHANISM AND REGULATION OF DNA END RESECTION IN EUKARYOTES.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 51. ISSUE 3. P. 195 -212 | 148 | 74% | 4 |
| 2 | THOMPSON, LH , (2012) RECOGNITION, SIGNALING, AND REPAIR OF DNA DOUBLE-STRAND BREAKS PRODUCED BY IONIZING RADIATION IN MAMMALIAN CELLS: THE MOLECULAR CHOREOGRAPHY.MUTATION RESEARCH-REVIEWS IN MUTATION RESEARCH. VOL. 751. ISSUE 2. P. 158 -246 | 320 | 26% | 151 |
| 3 | POLO, SE , JACKSON, SP , (2011) DYNAMICS OF DNA DAMAGE RESPONSE PROTEINS AT DNA BREAKS: A FOCUS ON PROTEIN MODIFICATIONS.GENES & DEVELOPMENT. VOL. 25. ISSUE 5. P. 409 -433 | 142 | 40% | 392 |
| 4 | GURSOY-YUZUGULLU, O , HOUSE, N , PRICE, BD , (2016) PATCHING BROKEN DNA: NUCLEOSOME DYNAMICS AND THE REPAIR OF DNA BREAKS.JOURNAL OF MOLECULAR BIOLOGY. VOL. 428. ISSUE 9. P. 1846 -1860 | 85 | 68% | 12 |
| 5 | PANIER, S , BOULTON, SJ , (2014) DOUBLE-STRAND BREAK REPAIR: 53BP1 COMES INTO FOCUS.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 15. ISSUE 1. P. 7 -18 | 75 | 65% | 155 |
| 6 | STRACKER, TH , PETRINI, JHJ , (2011) THE MRE11 COMPLEX: STARTING FROM THE ENDS.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 12. ISSUE 2. P. 90 -103 | 102 | 60% | 240 |
| 7 | RUPNIK, A , LOWNDES, NF , GRENON, M , (2010) MRN AND THE RACE TO THE BREAK.CHROMOSOMA. VOL. 119. ISSUE 2. P. 115 -135 | 133 | 72% | 32 |
| 8 | SYMINGTON, LS , GAUTIER, J , (2011) DOUBLE-STRAND BREAK END RESECTION AND REPAIR PATHWAY CHOICE.ANNUAL REVIEW OF GENETICS, VOL 45. VOL. 45. ISSUE . P. 247 -271 | 93 | 51% | 442 |
| 9 | DALEY, JM , SUNG, P , (2014) 53BP1, BRCA1, AND THE CHOICE BETWEEN RECOMBINATION AND END JOINING AT DNA DOUBLE-STRAND BREAKS.MOLECULAR AND CELLULAR BIOLOGY. VOL. 34. ISSUE 8. P. 1380-1388 | 83 | 82% | 41 |
| 10 | PAUDYAL, SC , YOU, ZS , (2016) SHARPENING THE ENDS FOR REPAIR: MECHANISMS AND REGULATION OF DNA RESECTION.ACTA BIOCHIMICA ET BIOPHYSICA SINICA. VOL. 48. ISSUE 7. P. 647 -657 | 121 | 57% | 1 |
Classes with closest relation at Level 1 |
| Rank | Class id | link |
|---|---|---|
| 1 | 13797 | GAMMA H2AX//GAMMA H2AX FOCI//H2AX |
| 2 | 3254 | DNA PK//NON HOMOLOGOUS END JOINING//DNA DEPENDENT PROTEIN KINASE |
| 3 | 2448 | CHK1//CHECKPOINT//RAD9 |
| 4 | 5917 | ATAXIA TELANGIECTASIA//ATM//ATM GENE |
| 5 | 4671 | RAD51//RAD52//HOMOLOGOUS RECOMBINATION |
| 6 | 5107 | BRCA1//BARD1//BRCA2 |
| 7 | 15199 | CHEK2//PALB2//CHK2 |
| 8 | 10345 | PARP INHIBITORS//OLAPARIB//SYNTHETIC LETHALITY |
| 9 | 21108 | FEN1//BRAUN S//CREAT INITIAT CELL CYCLE CONTROL |
| 10 | 1312 | TELOMERE//TELOMERASE//TRF2 |