Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
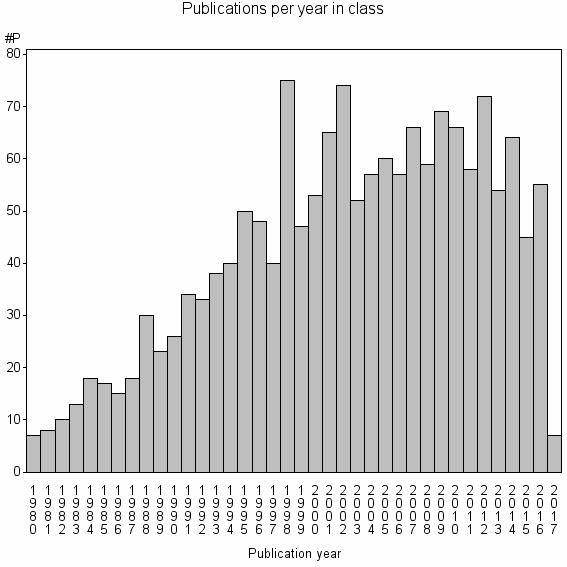
| 5439 | 1623 | 40.7 | 81% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 160 | 2 | WERNER SYNDROME//DNA POLYMERASE//DNA REPAIR | 22798 |
| 5439 | 1 | DNA POLYMERASE BETA//DNA POLYMERASE//DNA POLYMERASE LAMBDA | 1623 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | DNA POLYMERASE BETA | authKW | 577499 | 5% | 37% | 82 |
| 2 | DNA POLYMERASE | authKW | 473299 | 10% | 15% | 170 |
| 3 | DNA POLYMERASE LAMBDA | authKW | 248775 | 1% | 58% | 23 |
| 4 | DNA POLYMERASE MU | authKW | 112875 | 0% | 100% | 6 |
| 5 | DNA POLYMERASE I | authKW | 98827 | 1% | 31% | 17 |
| 6 | FAMILY B DNA POLYMERASE | authKW | 96748 | 0% | 86% | 6 |
| 7 | EXONUCLEASE ACTIVITY | authKW | 89627 | 1% | 53% | 9 |
| 8 | BASS MOL STRUCT BIOL | address | 85993 | 0% | 57% | 8 |
| 9 | TAQ DNA POLYMERASE | authKW | 75564 | 1% | 27% | 15 |
| 10 | FAMILY X | authKW | 75250 | 0% | 100% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 13362 | 73% | 0% | 1183 |
| 2 | Biophysics | 747 | 9% | 0% | 142 |
| 3 | Genetics & Heredity | 466 | 8% | 0% | 136 |
| 4 | Biotechnology & Applied Microbiology | 275 | 7% | 0% | 113 |
| 5 | Toxicology | 270 | 5% | 0% | 75 |
| 6 | Cell Biology | 231 | 8% | 0% | 132 |
| 7 | Biochemical Research Methods | 204 | 5% | 0% | 76 |
| 8 | Microbiology | 63 | 4% | 0% | 67 |
| 9 | Biology | 28 | 2% | 0% | 31 |
| 10 | Oncology | 12 | 4% | 0% | 61 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BASS MOL STRUCT BIOL | 85993 | 0% | 57% | 8 |
| 2 | STRUCT BIOL | 57189 | 9% | 2% | 142 |
| 3 | THER EUT RADIOL GENET | 54717 | 0% | 36% | 8 |
| 4 | JOSEPH GOTTSTEIN MEM CANC | 53866 | 1% | 22% | 13 |
| 5 | OHIO STATE BIOPHYS PROGRAM | 49468 | 1% | 24% | 11 |
| 6 | VET BIOCHEM MOL BIOL | 48465 | 2% | 8% | 32 |
| 7 | GRP ABIL GENET CANC | 42326 | 0% | 75% | 3 |
| 8 | GRP GENET ABIL CANC | 42326 | 0% | 75% | 3 |
| 9 | HARVARD MIT JOINT HLTH SCI TECHNOL | 37625 | 0% | 100% | 2 |
| 10 | SUGARY | 37625 | 0% | 100% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 17043 | 3% | 2% | 43 |
| 2 | BIOCHEMISTRY | 15761 | 13% | 0% | 212 |
| 3 | NUCLEIC ACIDS RESEARCH | 8806 | 8% | 0% | 137 |
| 4 | JOURNAL OF BIOLOGICAL CHEMISTRY | 4627 | 12% | 0% | 197 |
| 5 | JOURNAL OF MOLECULAR BIOLOGY | 3640 | 4% | 0% | 68 |
| 6 | EXTREMOPHILES | 1590 | 1% | 1% | 11 |
| 7 | BIOTECHNOLOGY EDUCATION | 1445 | 0% | 8% | 1 |
| 8 | STRUCTURE | 1403 | 1% | 0% | 17 |
| 9 | PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA | 1065 | 5% | 0% | 77 |
| 10 | MUTATION RESEARCH-DNA REPAIR | 956 | 0% | 1% | 5 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNA POLYMERASE BETA | 577499 | 5% | 37% | 82 | Search DNA+POLYMERASE+BETA | Search DNA+POLYMERASE+BETA |
| 2 | DNA POLYMERASE | 473299 | 10% | 15% | 170 | Search DNA+POLYMERASE | Search DNA+POLYMERASE |
| 3 | DNA POLYMERASE LAMBDA | 248775 | 1% | 58% | 23 | Search DNA+POLYMERASE+LAMBDA | Search DNA+POLYMERASE+LAMBDA |
| 4 | DNA POLYMERASE MU | 112875 | 0% | 100% | 6 | Search DNA+POLYMERASE+MU | Search DNA+POLYMERASE+MU |
| 5 | DNA POLYMERASE I | 98827 | 1% | 31% | 17 | Search DNA+POLYMERASE+I | Search DNA+POLYMERASE+I |
| 6 | FAMILY B DNA POLYMERASE | 96748 | 0% | 86% | 6 | Search FAMILY+B+DNA+POLYMERASE | Search FAMILY+B+DNA+POLYMERASE |
| 7 | EXONUCLEASE ACTIVITY | 89627 | 1% | 53% | 9 | Search EXONUCLEASE+ACTIVITY | Search EXONUCLEASE+ACTIVITY |
| 8 | TAQ DNA POLYMERASE | 75564 | 1% | 27% | 15 | Search TAQ+DNA+POLYMERASE | Search TAQ+DNA+POLYMERASE |
| 9 | FAMILY X | 75250 | 0% | 100% | 4 | Search FAMILY+X | Search FAMILY+X |
| 10 | THERMOSTABLE DNA POLYMERASE | 66880 | 0% | 44% | 8 | Search THERMOSTABLE+DNA+POLYMERASE | Search THERMOSTABLE+DNA+POLYMERASE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | YAMTICH, J , SWEASY, JB , (2010) DNA POLYMERASE FAMILY X: FUNCTION, STRUCTURE, AND CELLULAR ROLES.BIOCHIMICA ET BIOPHYSICA ACTA-PROTEINS AND PROTEOMICS. VOL. 1804. ISSUE 5. P. 1136-1150 | 118 | 70% | 53 |
| 2 | BEARD, WA , WILSON, SH , (2006) STRUCTURE AND MECHANISM OF DNA POLYMERASE BETA.CHEMICAL REVIEWS. VOL. 106. ISSUE 2. P. 361-382 | 110 | 72% | 178 |
| 3 | BEARD, WA , WILSON, SH , (2014) STRUCTURE AND MECHANISM OF DNA POLYMERASE BETA.BIOCHEMISTRY. VOL. 53. ISSUE 17. P. 2768 -2780 | 65 | 88% | 19 |
| 4 | BELOUSOVA, EA , LAVRIK, OI , (2015) DNA POLYMERASES BETA AND LAMBDA AND THEIR ROLES IN CELL.DNA REPAIR. VOL. 29. ISSUE . P. 112 -126 | 118 | 53% | 1 |
| 5 | MOON, AF , GARCIA-DIAZ, M , BATRA, VK , BEARD, WA , BEBENEK, K , KUNKEL, TA , WILSON, SH , PEDERSEN, LC , (2007) THE X FAMILY PORTRAIT: STRUCTURAL INSIGHTS INTO BIOLOGICAL FUNCTIONS OF X FAMILY POLYMERASES.DNA REPAIR. VOL. 6. ISSUE 12. P. 1709-1725 | 71 | 84% | 77 |
| 6 | MOSCATO, B , SWAIN, M , LORIA, JP , (2016) INDUCED FIT IN THE SELECTION OF CORRECT VERSUS INCORRECT NUCLEOTIDES BY DNA POLYMERASE BETA.BIOCHEMISTRY. VOL. 55. ISSUE 2. P. 382 -395 | 64 | 75% | 1 |
| 7 | BEBENEK, K , PEDERSEN, LC , KUNKEL, TA , (2014) STRUCTURE-FUNCTION STUDIES OF DNA POLYMERASE LAMBDA.BIOCHEMISTRY. VOL. 53. ISSUE 17. P. 2781-2792 | 62 | 79% | 11 |
| 8 | BEARD, WA , SHOCK, DD , BATRA, VK , PRASAD, R , WILSON, SH , (2014) SUBSTRATE-INDUCED DNA POLYMERASE BETA ACTIVATION.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 289. ISSUE 45. P. 31411 -31422 | 52 | 91% | 4 |
| 9 | BELOUSOVA, EA , LAVRIK, OI , (2010) DNA POLYMERASES BETA AND LAMBDA AND THEIR ROLES IN DNA REPLICATION AND REPAIR.MOLECULAR BIOLOGY. VOL. 44. ISSUE 6. P. 839-855 | 97 | 54% | 3 |
| 10 | TOWLE-WEICKSEL, JB , DALAL, S , SOHL, CD , DOUBLIE, S , ANDERSON, KS , SWEASY, JB , (2014) FLUORESCENCE RESONANCE ENERGY TRANSFER STUDIES OF DNA POLYMERASE BETA THE CRITICAL ROLE OF FINGERS DOMAIN MOVEMENTS AND A NOVEL NON-COVALENT STEP DURING NUCLEOTIDE SELECTION.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 289. ISSUE 23. P. 16541-16550 | 49 | 88% | 6 |
Classes with closest relation at Level 1 |