Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
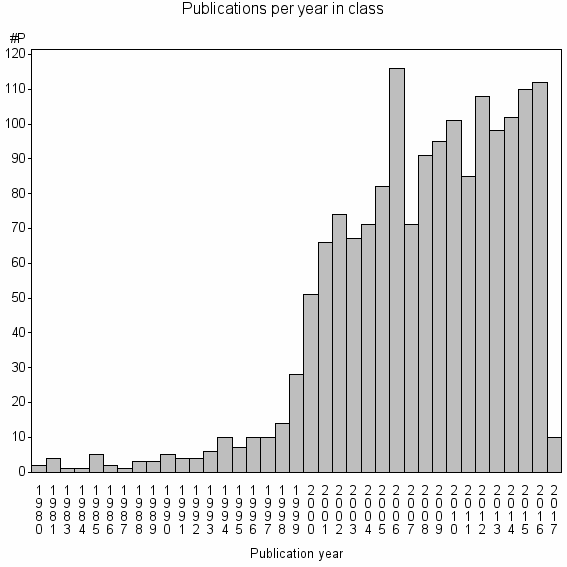
| 5392 | 1630 | 53.2 | 92% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 160 | 2 | WERNER SYNDROME//DNA POLYMERASE//DNA REPAIR | 22798 |
| 5392 | 1 | TRANSLESION SYNTHESIS//TRANSLESION DNA SYNTHESIS//DNA DAMAGE TOLERANCE | 1630 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | TRANSLESION SYNTHESIS | authKW | 1591357 | 9% | 60% | 141 |
| 2 | TRANSLESION DNA SYNTHESIS | authKW | 800165 | 4% | 60% | 71 |
| 3 | DNA DAMAGE TOLERANCE | authKW | 621805 | 3% | 74% | 45 |
| 4 | DNA POLYMERASE ETA | authKW | 619289 | 3% | 72% | 46 |
| 5 | REV1 | authKW | 527728 | 2% | 78% | 36 |
| 6 | Y FAMILY DNA POLYMERASE | authKW | 366337 | 2% | 67% | 29 |
| 7 | REV3 | authKW | 359638 | 1% | 80% | 24 |
| 8 | DNA POLYMERASE IOTA | authKW | 330419 | 1% | 84% | 21 |
| 9 | POST REPLICATION REPAIR | authKW | 302923 | 2% | 56% | 29 |
| 10 | POLYMERASE ETA | authKW | 270667 | 1% | 85% | 17 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 7741 | 57% | 0% | 924 |
| 2 | Genetics & Heredity | 6342 | 27% | 0% | 433 |
| 3 | Toxicology | 4858 | 17% | 0% | 274 |
| 4 | Cell Biology | 4760 | 29% | 0% | 465 |
| 5 | Biophysics | 237 | 5% | 0% | 89 |
| 6 | Oncology | 232 | 8% | 0% | 135 |
| 7 | Biotechnology & Applied Microbiology | 96 | 5% | 0% | 78 |
| 8 | Chemistry, Medicinal | 63 | 3% | 0% | 43 |
| 9 | Developmental Biology | 28 | 1% | 0% | 20 |
| 10 | Biology | 15 | 2% | 0% | 26 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENOM INTEGR | 240717 | 2% | 46% | 28 |
| 2 | SEALY MOL SCI | 206988 | 4% | 18% | 63 |
| 3 | SECT DNA REPLICAT REPAIR MUTAGENESIS | 184498 | 2% | 36% | 27 |
| 4 | GENOME ABIL SECT | 116939 | 1% | 39% | 16 |
| 5 | GENOME DAMAGE STABIL | 81493 | 3% | 9% | 50 |
| 6 | OHIO STATE BIOPHYS PROGRAM | 58620 | 1% | 26% | 12 |
| 7 | GENET INFORMAT ANAL | 56195 | 0% | 100% | 3 |
| 8 | RADIAT GENET | 55014 | 2% | 11% | 27 |
| 9 | MANIPAL EDU | 37463 | 0% | 100% | 2 |
| 10 | CANC GENOM IN IDUALIZED MED | 33715 | 0% | 60% | 3 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 193642 | 9% | 7% | 145 |
| 2 | NUCLEIC ACIDS RESEARCH | 8638 | 8% | 0% | 136 |
| 3 | MUTATION RESEARCH-DNA REPAIR | 7512 | 1% | 3% | 14 |
| 4 | MUTATION RESEARCH-FUNDAMENTAL AND MOLECULAR MECHANISMS OF MUTAGENESIS | 7255 | 2% | 1% | 39 |
| 5 | MOLECULAR CELL | 6890 | 3% | 1% | 45 |
| 6 | MOLECULAR AND CELLULAR BIOLOGY | 4151 | 4% | 0% | 71 |
| 7 | GENES TO CELLS | 3503 | 1% | 1% | 19 |
| 8 | CHEMICAL RESEARCH IN TOXICOLOGY | 2984 | 2% | 1% | 29 |
| 9 | JOURNAL OF BIOLOGICAL CHEMISTRY | 2458 | 9% | 0% | 146 |
| 10 | ENVIRONMENTAL AND MOLECULAR MUTAGENESIS | 2348 | 1% | 1% | 17 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TRANSLESION SYNTHESIS | 1591357 | 9% | 60% | 141 | Search TRANSLESION+SYNTHESIS | Search TRANSLESION+SYNTHESIS |
| 2 | TRANSLESION DNA SYNTHESIS | 800165 | 4% | 60% | 71 | Search TRANSLESION+DNA+SYNTHESIS | Search TRANSLESION+DNA+SYNTHESIS |
| 3 | DNA DAMAGE TOLERANCE | 621805 | 3% | 74% | 45 | Search DNA+DAMAGE+TOLERANCE | Search DNA+DAMAGE+TOLERANCE |
| 4 | DNA POLYMERASE ETA | 619289 | 3% | 72% | 46 | Search DNA+POLYMERASE+ETA | Search DNA+POLYMERASE+ETA |
| 5 | REV1 | 527728 | 2% | 78% | 36 | Search REV1 | Search REV1 |
| 6 | Y FAMILY DNA POLYMERASE | 366337 | 2% | 67% | 29 | Search Y+FAMILY+DNA+POLYMERASE | Search Y+FAMILY+DNA+POLYMERASE |
| 7 | REV3 | 359638 | 1% | 80% | 24 | Search REV3 | Search REV3 |
| 8 | DNA POLYMERASE IOTA | 330419 | 1% | 84% | 21 | Search DNA+POLYMERASE+IOTA | Search DNA+POLYMERASE+IOTA |
| 9 | POST REPLICATION REPAIR | 302923 | 2% | 56% | 29 | Search POST+REPLICATION+REPAIR | Search POST+REPLICATION+REPAIR |
| 10 | POLYMERASE ETA | 270667 | 1% | 85% | 17 | Search POLYMERASE+ETA | Search POLYMERASE+ETA |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | WATERS, LS , MINESINGER, BK , WILTROUT, ME , D'SOUZA, S , WOODRUFF, RV , WALKER, GC , (2009) EUKARYOTIC TRANSLESION POLYMERASES AND THEIR ROLES AND REGULATION IN DNA DAMAGE TOLERANCE.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 73. ISSUE 1. P. 134-+ | 208 | 77% | 241 |
| 2 | SALE, JE , (2013) TRANSLESION DNA SYNTHESIS AND MUTAGENESIS IN EUKARYOTES.COLD SPRING HARBOR PERSPECTIVES IN BIOLOGY. VOL. 5. ISSUE 3. P. - | 178 | 71% | 32 |
| 3 | GUO, CX , KOSAREK-STANCEL, J , TANG, TS , FRIEDBERG, EC , (2009) Y-FAMILY DNA POLYMERASES IN MAMMALIAN CELLS.CELLULAR AND MOLECULAR LIFE SCIENCES. VOL. 66. ISSUE 14. P. 2363 -2381 | 156 | 85% | 79 |
| 4 | SHARMA, S , HELCHOWSKI, CM , CANMAN, CE , (2013) THE ROLES OF DNA POLYMERASE ZETA AND THE Y FAMILY DNA POLYMERASES IN PROMOTING OR PREVENTING GENOME INSTABILITY.MUTATION RESEARCH-FUNDAMENTAL AND MOLECULAR MECHANISMS OF MUTAGENESIS. VOL. 743. ISSUE . P. 97 -110 | 160 | 68% | 24 |
| 5 | SALE, JE , LEHMANN, AR , WOODGATE, R , (2012) Y-FAMILY DNA POLYMERASES AND THEIR ROLE IN TOLERANCE OF CELLULAR DNA DAMAGE.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 13. ISSUE 3. P. 141 -152 | 92 | 77% | 217 |
| 6 | LANGE, SS , TAKATA, K , WOOD, RD , (2011) DNA POLYMERASES AND CANCER.NATURE REVIEWS CANCER. VOL. 11. ISSUE 2. P. 96-110 | 124 | 65% | 196 |
| 7 | JANSEN, JG , TSAALBI-SHTYLIK, A , DE WIND, N , (2015) ROLES OF MUTAGENIC TRANSLESION SYNTHESIS IN MAMMALIAN GENOME STABILITY, HEALTH AND DISEASE.DNA REPAIR. VOL. 29. ISSUE . P. 56 -64 | 115 | 83% | 9 |
| 8 | KORZHNEV, DM , HADDEN, MK , (2016) TARGETING THE TRANSLESION SYNTHESIS PATHWAY FOR THE DEVELOPMENT OF ANTI-CANCER CHEMOTHERAPEUTICS.JOURNAL OF MEDICINAL CHEMISTRY. VOL. 59. ISSUE 20. P. 9321 -9336 | 126 | 72% | 1 |
| 9 | MAKAROVA, AV , BURGERS, PM , (2015) EUKARYOTIC DNA POLYMERASE ZETA.DNA REPAIR. VOL. 29. ISSUE . P. 47 -55 | 102 | 74% | 14 |
| 10 | TOMCZYK, P , SYNOWIEC, E , WYSOKINSKI, D , WOZNIAK, K , (2016) EUKARYOTIC TLS POLYMERASES.POSTEPY HIGIENY I MEDYCYNY DOSWIADCZALNEJ. VOL. 70. ISSUE . P. 522 -533 | 105 | 86% | 0 |
Classes with closest relation at Level 1 |