Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
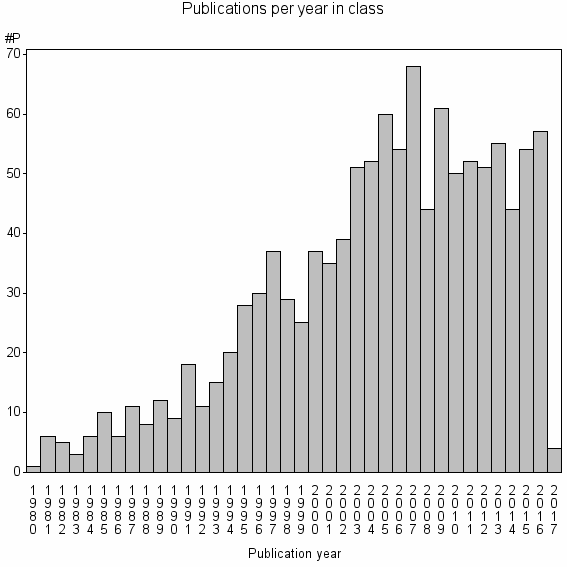
| 9408 | 1158 | 39.1 | 79% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 5 | 4 | CHEMISTRY, ORGANIC//CHEMISTRY, INORGANIC & NUCLEAR//CHEMISTRY, MULTIDISCIPLINARY | 1745167 |
| 183 | 3 | APTAMER//G QUADRUPLEX//RIBOZYME | 56307 |
| 389 | 2 | NUCLEOSIDES NUCLEOTIDES & NUCLEIC ACIDS//OLIGONUCLEOTIDES//PEPTIDE NUCLEIC ACID | 16949 |
| 9408 | 1 | METAL MEDIATED BASE PAIRS//UNNATURAL BASE PAIRS//METALLO BASE PAIR | 1158 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | METAL MEDIATED BASE PAIRS | authKW | 660161 | 2% | 96% | 26 |
| 2 | UNNATURAL BASE PAIRS | authKW | 303996 | 1% | 82% | 14 |
| 3 | METALLO BASE PAIR | authKW | 158205 | 1% | 100% | 6 |
| 4 | PROT NUCLE ACID | address | 129196 | 1% | 70% | 7 |
| 5 | C NUCLEOSIDES | authKW | 122031 | 2% | 19% | 25 |
| 6 | AMERSHAM S | address | 118645 | 1% | 50% | 9 |
| 7 | MOL TRANSFORMAT | address | 112399 | 1% | 47% | 9 |
| 8 | GENETIC ALPHABET | authKW | 94167 | 0% | 71% | 5 |
| 9 | ARTIFICIAL DNA | authKW | 92279 | 1% | 50% | 7 |
| 10 | HYDROPHOBIC BASES | authKW | 84374 | 0% | 80% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Chemistry, Organic | 2580 | 22% | 0% | 258 |
| 2 | Chemistry, Multidisciplinary | 2352 | 30% | 0% | 353 |
| 3 | Biochemistry & Molecular Biology | 1565 | 33% | 0% | 380 |
| 4 | Chemistry, Medicinal | 202 | 5% | 0% | 54 |
| 5 | Chemistry, Inorganic & Nuclear | 199 | 6% | 0% | 66 |
| 6 | Chemistry, Physical | 56 | 8% | 0% | 93 |
| 7 | Physics, Atomic, Molecular & Chemical | 51 | 4% | 0% | 46 |
| 8 | Crystallography | 35 | 2% | 0% | 25 |
| 9 | Biophysics | 27 | 3% | 0% | 34 |
| 10 | Biochemical Research Methods | 10 | 2% | 0% | 23 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROT NUCLE ACID | 129196 | 1% | 70% | 7 |
| 2 | AMERSHAM S | 118645 | 1% | 50% | 9 |
| 3 | MOL TRANSFORMAT | 112399 | 1% | 47% | 9 |
| 4 | SSBC | 70311 | 0% | 67% | 4 |
| 5 | WESTHEIMER SCI TECHNOL | 70311 | 0% | 67% | 4 |
| 6 | MODELING SIMULAT CHEM | 56191 | 1% | 24% | 9 |
| 7 | COMPUTAC BIOL PROGRAM | 52735 | 0% | 100% | 2 |
| 8 | FELLOW GRP | 52735 | 0% | 100% | 2 |
| 9 | CHEM NEUER MAT | 50690 | 1% | 19% | 10 |
| 10 | YOKOYAMA CYTOLOG PROJECT | 47453 | 1% | 30% | 6 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NUCLEOSIDES NUCLEOTIDES & NUCLEIC ACIDS | 29267 | 6% | 2% | 72 |
| 2 | NUCLEIC ACIDS RESEARCH | 4617 | 7% | 0% | 84 |
| 3 | JOURNAL OF THE AMERICAN CHEMICAL SOCIETY | 3468 | 10% | 0% | 112 |
| 4 | NUCLEOSIDES & NUCLEOTIDES | 2522 | 1% | 1% | 8 |
| 5 | CHEMBIOCHEM | 2110 | 2% | 0% | 19 |
| 6 | ANGEWANDTE CHEMIE-INTERNATIONAL EDITION | 1839 | 5% | 0% | 53 |
| 7 | HELVETICA CHIMICA ACTA | 1180 | 2% | 0% | 20 |
| 8 | CHEMISTRY-A EUROPEAN JOURNAL | 1180 | 3% | 0% | 33 |
| 9 | JOURNAL OF ORGANIC CHEMISTRY | 1167 | 4% | 0% | 49 |
| 10 | BIOCHEMISTRY | 849 | 4% | 0% | 43 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | METAL MEDIATED BASE PAIRS | 660161 | 2% | 96% | 26 | Search METAL+MEDIATED+BASE+PAIRS | Search METAL+MEDIATED+BASE+PAIRS |
| 2 | UNNATURAL BASE PAIRS | 303996 | 1% | 82% | 14 | Search UNNATURAL+BASE+PAIRS | Search UNNATURAL+BASE+PAIRS |
| 3 | METALLO BASE PAIR | 158205 | 1% | 100% | 6 | Search METALLO+BASE+PAIR | Search METALLO+BASE+PAIR |
| 4 | C NUCLEOSIDES | 122031 | 2% | 19% | 25 | Search C+NUCLEOSIDES | Search C+NUCLEOSIDES |
| 5 | GENETIC ALPHABET | 94167 | 0% | 71% | 5 | Search GENETIC+ALPHABET | Search GENETIC+ALPHABET |
| 6 | ARTIFICIAL DNA | 92279 | 1% | 50% | 7 | Search ARTIFICIAL+DNA | Search ARTIFICIAL+DNA |
| 7 | HYDROPHOBIC BASES | 84374 | 0% | 80% | 4 | Search HYDROPHOBIC+BASES | Search HYDROPHOBIC+BASES |
| 8 | ARTIFICIALLY EXPANDED GENETIC INFORMATION SYSTEMS | 79102 | 0% | 100% | 3 | Search ARTIFICIALLY+EXPANDED+GENETIC+INFORMATION+SYSTEMS | Search ARTIFICIALLY+EXPANDED+GENETIC+INFORMATION+SYSTEMS |
| 9 | EXPANDED DNA | 79102 | 0% | 100% | 3 | Search EXPANDED+DNA | Search EXPANDED+DNA |
| 10 | METALLO DNA | 79102 | 0% | 100% | 3 | Search METALLO+DNA | Search METALLO+DNA |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | STAMBASKY, J , HOCEK, M , KOCOVSKY, P , (2009) C-NUCLEOSIDES: SYNTHETIC STRATEGIES AND BIOLOGICAL APPLICATIONS.CHEMICAL REVIEWS. VOL. 109. ISSUE 12. P. 6729 -6764 | 178 | 49% | 108 |
| 2 | MALYSHEV, DA , ROMESBERG, FE , (2015) THE EXPANDED GENETIC ALPHABET.ANGEWANDTE CHEMIE-INTERNATIONAL EDITION. VOL. 54. ISSUE 41. P. 11930 -11944 | 95 | 52% | 13 |
| 3 | HIRAO, I , KIMOTO, M , YAMASHIGE, R , (2012) NATURAL VERSUS ARTIFICIAL CREATION OF BASE PAIRS IN DNA: ORIGIN OF NUCLEOBASES FROM THE PERSPECTIVES OF UNNATURAL BASE PAIR STUDIES.ACCOUNTS OF CHEMICAL RESEARCH. VOL. 45. ISSUE 12. P. 2055-2065 | 51 | 89% | 48 |
| 4 | TANAKA, Y , KONDO, J , SYCHROVSKY, V , SEBERA, J , DAIRAKU, T , SANEYOSHI, H , URATA, H , TORIGOE, H , ONO, A , (2015) STRUCTURES, PHYSICOCHEMICAL PROPERTIES, AND APPLICATIONS OF T-HG-II-T, C-AG-I-C, AND OTHER METALLO-BASE-PAIRS.CHEMICAL COMMUNICATIONS. VOL. 51. ISSUE 98. P. 17343 -17360 | 73 | 54% | 11 |
| 5 | CHELLISERRYKATTIL, J , LU, H , LEE, AHF , KOOL, ET , (2008) POLYMERASE AMPLIFICATION, CLONING, AND GENE EXPRESSION OF BENZO-HOMOLOGOUS "YDNA" BASE PAIRS.CHEMBIOCHEM. VOL. 9. ISSUE 18. P. 2976-2980 | 54 | 96% | 14 |
| 6 | WOJCIECHOWSKI, F , LEUMANN, CJ , (2011) ALTERNATIVE DNA BASE-PAIRS: FROM EFFORTS TO EXPAND THE GENETIC CODE TO POTENTIAL MATERIAL APPLICATIONS.CHEMICAL SOCIETY REVIEWS. VOL. 40. ISSUE 12. P. 5669 -5679 | 56 | 75% | 41 |
| 7 | KIMOTO, M , HIKIDA, Y , HIRAO, I , (2013) SITE-SPECIFIC FUNCTIONAL LABELING OF NUCLEIC ACIDS BY IN VITRO REPLICATION AND TRANSCRIPTION USING UNNATURAL BASE PAIR SYSTEMS.ISRAEL JOURNAL OF CHEMISTRY. VOL. 53. ISSUE 6-7. P. 450-468 | 73 | 61% | 5 |
| 8 | LIANG, F , LIU, YZ , ZHANG, PM , (2013) UNIVERSAL BASE ANALOGUES AND THEIR APPLICATIONS IN DNA SEQUENCING TECHNOLOGY.RSC ADVANCES. VOL. 3. ISSUE 35. P. 14910-14928 | 83 | 51% | 6 |
| 9 | KIMOTO, M , COX, RS , HIRAO, I , (2011) UNNATURAL BASE PAIR SYSTEMS FOR SENSING AND DIAGNOSTIC APPLICATIONS.EXPERT REVIEW OF MOLECULAR DIAGNOSTICS. VOL. 11. ISSUE 3. P. 321 -331 | 57 | 74% | 19 |
| 10 | HIRAO, I , KIMOTO, M , (2012) UNNATURAL BASE PAIR SYSTEMS TOWARD THE EXPANSION OF THE GENETIC ALPHABET IN THE CENTRAL DOGMA.PROCEEDINGS OF THE JAPAN ACADEMY SERIES B-PHYSICAL AND BIOLOGICAL SCIENCES. VOL. 88. ISSUE 7. P. 345 -367 | 55 | 71% | 13 |
Classes with closest relation at Level 1 |