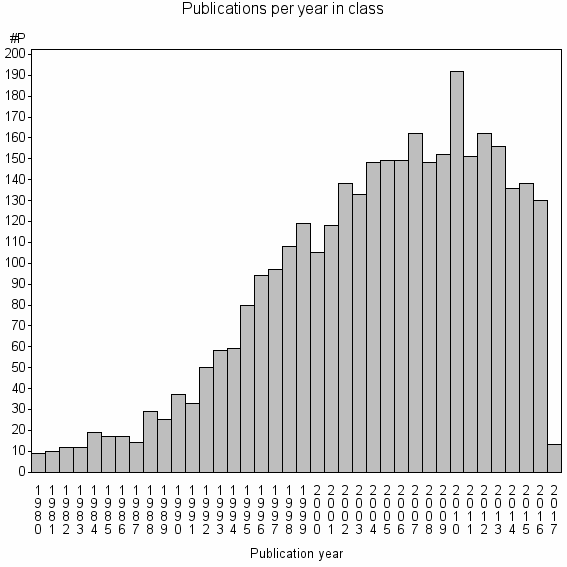
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 2682 | 3379 | 50.8 | 90% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 71 | 3 | GENETICS & HEREDITY//CHROMATIN//CELL BIOLOGY | 83302 |
| 2682 | 2 | DNA REPLICATION//GEMININ//CDC6 | 3379 |
| 577 | 1 | DNA REPLICATION//GEMININ//CDC6 | 3286 |
| 35681 | 1 | FKBP6//AGGREGATE OF RIBOSOME//AS MUTANT | 93 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | DNA REPLICATION | authKW | 1022606 | 17% | 20% | 570 |
| 2 | GEMININ | authKW | 623442 | 3% | 76% | 91 |
| 3 | CDC6 | authKW | 574288 | 2% | 76% | 84 |
| 4 | CDT1 | authKW | 517827 | 2% | 87% | 66 |
| 5 | ORIGIN RECOGNITION COMPLEX | authKW | 429280 | 2% | 85% | 56 |
| 6 | REPLICATION ORIGIN | authKW | 399920 | 3% | 42% | 105 |
| 7 | CDC7 | authKW | 267681 | 1% | 74% | 40 |
| 8 | MCM PROTEINS | authKW | 257378 | 1% | 81% | 35 |
| 9 | MCM | authKW | 256210 | 2% | 37% | 77 |
| 10 | ORC | authKW | 242872 | 3% | 31% | 86 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Cell Biology | 30710 | 49% | 0% | 1640 |
| 2 | Biochemistry & Molecular Biology | 15984 | 57% | 0% | 1912 |
| 3 | Genetics & Heredity | 5327 | 17% | 0% | 591 |
| 4 | Developmental Biology | 1789 | 5% | 0% | 168 |
| 5 | Oncology | 292 | 7% | 0% | 236 |
| 6 | Biophysics | 197 | 4% | 0% | 132 |
| 7 | Biology | 113 | 2% | 0% | 80 |
| 8 | Biotechnology & Applied Microbiology | 80 | 4% | 0% | 123 |
| 9 | Biochemical Research Methods | 48 | 2% | 0% | 77 |
| 10 | Mycology | 39 | 1% | 0% | 20 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPLICAT GRP | 165161 | 1% | 63% | 29 |
| 2 | CHROMOSOME REPLICAT | 119577 | 0% | 88% | 15 |
| 3 | CHROMOSOME REPLICAT GRP | 91100 | 0% | 92% | 11 |
| 4 | GENOME DYNAM PROJECT | 79664 | 1% | 42% | 21 |
| 5 | CLARE HALL S | 53011 | 2% | 9% | 63 |
| 6 | CRC CHROMOSOME REPLICAT GRP | 37644 | 0% | 83% | 5 |
| 7 | CANC CELL UNIT | 33149 | 1% | 14% | 26 |
| 8 | CANC UK CHROMOSOME REPLICAT GRP | 27105 | 0% | 100% | 3 |
| 9 | FUNCT ORG GENOME GRP | 27105 | 0% | 100% | 3 |
| 10 | HLTH MED MICROBIOL | 27105 | 0% | 100% | 3 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR AND CELLULAR BIOLOGY | 17863 | 6% | 1% | 211 |
| 2 | GENES & DEVELOPMENT | 13503 | 3% | 1% | 106 |
| 3 | COLD SPRING HARBOR SYMPOSIA ON QUANTITATIVE BIOLOGY SERIES | 12811 | 2% | 2% | 84 |
| 4 | MOLECULAR CELL | 7331 | 2% | 1% | 67 |
| 5 | GENES TO CELLS | 6756 | 1% | 2% | 38 |
| 6 | EMBO JOURNAL | 6238 | 3% | 1% | 110 |
| 7 | NUCLEIC ACIDS RESEARCH | 6149 | 5% | 0% | 167 |
| 8 | MOLECULAR BIOLOGY OF THE CELL | 5672 | 2% | 1% | 73 |
| 9 | CHROMOSOMA | 5362 | 1% | 2% | 40 |
| 10 | CELL DIVISION | 5286 | 0% | 5% | 11 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNA REPLICATION | 1022606 | 17% | 20% | 570 | Search DNA+REPLICATION | Search DNA+REPLICATION |
| 2 | GEMININ | 623442 | 3% | 76% | 91 | Search GEMININ | Search GEMININ |
| 3 | CDC6 | 574288 | 2% | 76% | 84 | Search CDC6 | Search CDC6 |
| 4 | CDT1 | 517827 | 2% | 87% | 66 | Search CDT1 | Search CDT1 |
| 5 | ORIGIN RECOGNITION COMPLEX | 429280 | 2% | 85% | 56 | Search ORIGIN+RECOGNITION+COMPLEX | Search ORIGIN+RECOGNITION+COMPLEX |
| 6 | REPLICATION ORIGIN | 399920 | 3% | 42% | 105 | Search REPLICATION+ORIGIN | Search REPLICATION+ORIGIN |
| 7 | CDC7 | 267681 | 1% | 74% | 40 | Search CDC7 | Search CDC7 |
| 8 | MCM PROTEINS | 257378 | 1% | 81% | 35 | Search MCM+PROTEINS | Search MCM+PROTEINS |
| 9 | MCM | 256210 | 2% | 37% | 77 | Search MCM | Search MCM |
| 10 | ORC | 242872 | 3% | 31% | 86 | Search ORC | Search ORC |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | BELL, SP , DUTTA, A , (2002) DNA REPLICATION IN EUKARYOTIC CELLS.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 71. ISSUE . P. 333-374 | 207 | 89% | 1112 |
| 2 | FORSBURG, SL , (2004) EUKARYOTIC MCM PROTEINS: BEYOND REPLICATION INITIATION.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 68. ISSUE 1. P. 109-+ | 280 | 81% | 291 |
| 3 | MASAI, H , MATSUMOTO, S , YOU, ZY , YOSHIZAWA-SUGATA, N , ODA, M , (2010) EUKARYOTIC CHROMOSOME DNA REPLICATION: WHERE, WHEN AND HOW?.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 79. VOL. 79. ISSUE . P. 89 -130 | 225 | 73% | 183 |
| 4 | SIDDIQUI, K , ON, KF , DIFFLEY, JFX , (2013) REGULATING DNA REPLICATION IN EUKARYA.COLD SPRING HARBOR PERSPECTIVES IN BIOLOGY. VOL. 5. ISSUE 9. P. - | 188 | 78% | 37 |
| 5 | ARIAS, EE , WALTER, JC , (2007) STRENGTH IN NUMBERS: PREVENTING REREPLICATION VIA MULTIPLE MECHANISMS IN EUKARYOTIC CELLS.GENES & DEVELOPMENT. VOL. 21. ISSUE 5. P. 497 -518 | 164 | 87% | 233 |
| 6 | HYRIEN, O , (2015) PEAKS CLOAKED IN THE MIST: THE LANDSCAPE OF MAMMALIAN REPLICATION ORIGINS.JOURNAL OF CELL BIOLOGY. VOL. 208. ISSUE 2. P. 147 -160 | 143 | 87% | 16 |
| 7 | SCLAFANI, RA , HOLZEN, TM , (2007) CELL CYCLE REGULATION OF DNA REPLICATION.ANNUAL REVIEW OF GENETICS. VOL. 41. ISSUE . P. 237-280 | 197 | 68% | 215 |
| 8 | FRAGKOS, M , GANIER, O , COULOMBE, P , MECHALI, M , (2015) DNA REPLICATION ORIGIN ACTIVATION IN SPACE AND TIME.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 16. ISSUE 6. P. 360 -374 | 138 | 75% | 23 |
| 9 | MECHALI, M , (2010) EUKARYOTIC DNA REPLICATION ORIGINS: MANY CHOICES FOR APPROPRIATE ANSWERS.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 11. ISSUE 10. P. 728 -738 | 128 | 83% | 185 |
| 10 | BOCHMAN, ML , SCHWACHA, A , (2009) THE MCM COMPLEX: UNWINDING THE MECHANISM OF A REPLICATIVE HELICASE.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 73. ISSUE 4. P. 652 -683 | 181 | 69% | 112 |
Classes with closest relation at Level 2 |