Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
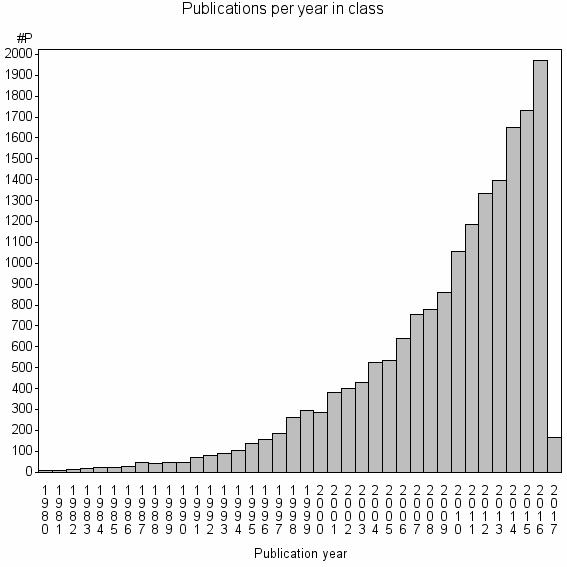
| 343 | 17754 | 49.7 | 92% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | EZH2 | authKW | 397786 | 2% | 65% | 356 |
| 2 | CHROMATIN | authKW | 349649 | 6% | 20% | 1026 |
| 3 | POLYCOMB | authKW | 262319 | 2% | 57% | 270 |
| 4 | RHABDOID TUMOR | authKW | 219318 | 1% | 76% | 168 |
| 5 | BROMODOMAIN | authKW | 189975 | 1% | 75% | 148 |
| 6 | SWI SNF | authKW | 182159 | 1% | 63% | 168 |
| 7 | ATYPICAL TERATOID RHABDOID TUMOR | authKW | 176440 | 1% | 81% | 127 |
| 8 | HISTONE | authKW | 176248 | 3% | 22% | 475 |
| 9 | HISTONE DEMETHYLASE | authKW | 173992 | 1% | 71% | 142 |
| 10 | NUCLEOSOME | authKW | 152331 | 2% | 29% | 306 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Cell Biology | 71098 | 33% | 1% | 5860 |
| 2 | Biochemistry & Molecular Biology | 56836 | 48% | 1% | 8438 |
| 3 | Genetics & Heredity | 25880 | 17% | 1% | 2997 |
| 4 | Oncology | 12030 | 15% | 0% | 2701 |
| 5 | Developmental Biology | 9409 | 5% | 1% | 883 |
| 6 | Biochemical Research Methods | 4373 | 6% | 0% | 1090 |
| 7 | Biophysics | 2881 | 6% | 0% | 1011 |
| 8 | Biotechnology & Applied Microbiology | 2316 | 6% | 0% | 1120 |
| 9 | Mathematical & Computational Biology | 1909 | 2% | 0% | 393 |
| 10 | Pathology | 1863 | 4% | 0% | 638 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | STRUCT GENOM CONSORTIUM | 90949 | 1% | 25% | 212 |
| 2 | CHROMATIN BIOL | 35599 | 0% | 36% | 58 |
| 3 | CHROMATIN BIOL EPIGENET | 32295 | 0% | 47% | 40 |
| 4 | BIOCHEM MOL BIOL | 25818 | 6% | 1% | 1108 |
| 5 | ADOLF BUTENANDT | 25640 | 1% | 16% | 93 |
| 6 | EPIGENET PROGRAM | 25526 | 0% | 34% | 44 |
| 7 | EPIGENET | 24157 | 1% | 13% | 107 |
| 8 | EPINOVA DPU | 22345 | 0% | 52% | 25 |
| 9 | INTEGRAT CHEM BIOL DRUG DISCOVERY | 20180 | 0% | 37% | 32 |
| 10 | CHROMATIN GENE EXP S SECT | 19616 | 0% | 57% | 20 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR CELL | 47356 | 2% | 7% | 390 |
| 2 | EPIGENETICS & CHROMATIN | 45911 | 0% | 30% | 88 |
| 3 | MOLECULAR AND CELLULAR BIOLOGY | 37449 | 4% | 3% | 705 |
| 4 | GENES & DEVELOPMENT | 24340 | 2% | 4% | 328 |
| 5 | GENOME RESEARCH | 19937 | 1% | 9% | 127 |
| 6 | EPIGENOMICS | 16085 | 0% | 14% | 66 |
| 7 | NATURE STRUCTURAL & MOLECULAR BIOLOGY | 14903 | 1% | 6% | 138 |
| 8 | BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS | 13422 | 1% | 9% | 91 |
| 9 | NUCLEIC ACIDS RESEARCH | 11473 | 3% | 1% | 532 |
| 10 | CELL | 9851 | 2% | 2% | 300 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | EZH2 | 397786 | 2% | 65% | 356 | Search EZH2 | Search EZH2 |
| 2 | CHROMATIN | 349649 | 6% | 20% | 1026 | Search CHROMATIN | Search CHROMATIN |
| 3 | POLYCOMB | 262319 | 2% | 57% | 270 | Search POLYCOMB | Search POLYCOMB |
| 4 | RHABDOID TUMOR | 219318 | 1% | 76% | 168 | Search RHABDOID+TUMOR | Search RHABDOID+TUMOR |
| 5 | BROMODOMAIN | 189975 | 1% | 75% | 148 | Search BROMODOMAIN | Search BROMODOMAIN |
| 6 | SWI SNF | 182159 | 1% | 63% | 168 | Search SWI+SNF | Search SWI+SNF |
| 7 | ATYPICAL TERATOID RHABDOID TUMOR | 176440 | 1% | 81% | 127 | Search ATYPICAL+TERATOID+RHABDOID+TUMOR | Search ATYPICAL+TERATOID+RHABDOID+TUMOR |
| 8 | HISTONE | 176248 | 3% | 22% | 475 | Search HISTONE | Search HISTONE |
| 9 | HISTONE DEMETHYLASE | 173992 | 1% | 71% | 142 | Search HISTONE+DEMETHYLASE | Search HISTONE+DEMETHYLASE |
| 10 | NUCLEOSOME | 152331 | 2% | 29% | 306 | Search NUCLEOSOME | Search NUCLEOSOME |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | RANDO, OJ , WINSTON, F , (2012) CHROMATIN AND TRANSCRIPTION IN YEAST.GENETICS. VOL. 190. ISSUE 2. P. 351-387 | 392 | 73% | 80 |
| 2 | MCGRATH, J , TROJER, P , (2015) TARGETING HISTONE LYSINE METHYLATION IN CANCER.PHARMACOLOGY & THERAPEUTICS. VOL. 150. ISSUE . P. 1 -22 | 217 | 67% | 39 |
| 3 | VENKATESH, S , WORKMAN, JL , (2015) HISTONE EXCHANGE, CHROMATIN STRUCTURE AND THE REGULATION OF TRANSCRIPTION.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 16. ISSUE 3. P. 178 -189 | 129 | 83% | 92 |
| 4 | KOOISTRA, SM , HELIN, K , (2012) MOLECULAR MECHANISMS AND POTENTIAL FUNCTIONS OF HISTONE DEMETHYLASES.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 13. ISSUE 5. P. 297-311 | 162 | 82% | 233 |
| 5 | BOWMAN, GD , POIRIER, MG , (2015) POST-TRANSLATIONAL MODIFICATIONS OF HISTONES THAT INFLUENCE NUCLEOSOME DYNAMICS.CHEMICAL REVIEWS. VOL. 115. ISSUE 6. P. 2274 -2295 | 234 | 69% | 22 |
| 6 | HU, H , QIAN, K , HO, MC , ZHENG, YG , (2016) SMALL MOLECULE INHIBITORS OF PROTEIN ARGININE METHYLTRANSFERASES.EXPERT OPINION ON INVESTIGATIONAL DRUGS. VOL. 25. ISSUE 3. P. 335 -358 | 175 | 81% | 6 |
| 7 | SIMON, JA , KINGSTON, RE , (2013) OCCUPYING CHROMATIN: POLYCOMB MECHANISMS FOR GETTING TO GENOMIC TARGETS, STOPPING TRANSCRIPTIONAL TRAFFIC, AND STAYING PUT.MOLECULAR CELL. VOL. 49. ISSUE 5. P. 808 -824 | 126 | 82% | 244 |
| 8 | LI, B , CAREY, M , WORKMAN, JL , (2007) THE ROLE OF CHROMATIN DURING TRANSCRIPTION.CELL. VOL. 128. ISSUE 4. P. 707 -719 | 101 | 81% | 1694 |
| 9 | WANG, W , QIN, JJ , VORUGANTI, S , NAG, S , ZHOU, JW , ZHANG, RW , (2015) POLYCOMB GROUP (PCG) PROTEINS AND HUMAN CANCERS: MULTIFACETED FUNCTIONS AND THERAPEUTIC IMPLICATIONS.MEDICINAL RESEARCH REVIEWS. VOL. 35. ISSUE 6. P. 1220 -1267 | 242 | 77% | 3 |
| 10 | KIM, KH , ROBERTS, CWM , (2016) TARGETING EZH2 IN CANCER.NATURE MEDICINE. VOL. 22. ISSUE 2. P. 128 -134 | 92 | 79% | 36 |
Classes with closest relation at Level 2 |