Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
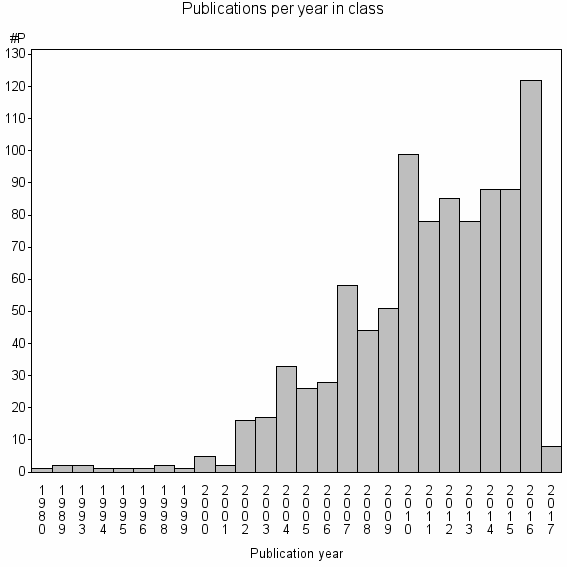
| 12023 | 937 | 50.9 | 92% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 71 | 3 | GENETICS & HEREDITY//CHROMATIN//CELL BIOLOGY | 83302 |
| 343 | 2 | EZH2//CHROMATIN//POLYCOMB | 17754 |
| 12023 | 1 | SET8//HISTONE//PR SET7 | 937 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | SET8 | authKW | 379185 | 2% | 73% | 16 |
| 2 | HISTONE | authKW | 243551 | 14% | 6% | 128 |
| 3 | PR SET7 | authKW | 232758 | 1% | 71% | 10 |
| 4 | CHROMATIN BIOL | address | 193259 | 3% | 19% | 31 |
| 5 | SPINDLIN1 | authKW | 177415 | 1% | 78% | 7 |
| 6 | ULTZ 415 | address | 162935 | 1% | 100% | 5 |
| 7 | HISTONE CODE | authKW | 133464 | 3% | 16% | 26 |
| 8 | EPIGENET PROGRAM | address | 132566 | 2% | 18% | 23 |
| 9 | SETD8 | authKW | 130344 | 1% | 67% | 6 |
| 10 | MBT DOMAIN | authKW | 104277 | 0% | 80% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 3635 | 52% | 0% | 485 |
| 2 | Biochemical Research Methods | 3115 | 20% | 0% | 185 |
| 3 | Cell Biology | 2322 | 26% | 0% | 248 |
| 4 | Genetics & Heredity | 385 | 10% | 0% | 91 |
| 5 | Biophysics | 180 | 6% | 0% | 57 |
| 6 | Developmental Biology | 100 | 2% | 0% | 23 |
| 7 | Chemistry, Analytical | 82 | 6% | 0% | 52 |
| 8 | Oncology | 76 | 7% | 0% | 64 |
| 9 | Chemistry, Medicinal | 62 | 3% | 0% | 30 |
| 10 | Spectroscopy | 34 | 2% | 0% | 22 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CHROMATIN BIOL | 193259 | 3% | 19% | 31 |
| 2 | ULTZ 415 | 162935 | 1% | 100% | 5 |
| 3 | EPIGENET PROGRAM | 132566 | 2% | 18% | 23 |
| 4 | TOP DOWN PROTEOM | 104277 | 0% | 80% | 4 |
| 5 | HISTONE MODIFICAT GRP | 84031 | 1% | 37% | 7 |
| 6 | INTEGRAT CHEM BIOL DRUG DISCOVERY | 73391 | 1% | 16% | 14 |
| 7 | CHROMATIN BIOCHEM | 72400 | 1% | 22% | 10 |
| 8 | PROGRAMMA DOTTORATO RIC SCI FARMACO | 65174 | 0% | 100% | 2 |
| 9 | CHROMATIN BIOL EPIGENET | 55185 | 1% | 14% | 12 |
| 10 | NEW JERSEY CANC | 43448 | 0% | 67% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR & CELLULAR PROTEOMICS | 10123 | 3% | 1% | 31 |
| 2 | EPIGENOMICS | 7034 | 1% | 2% | 10 |
| 3 | EPIGENETICS & CHROMATIN | 5511 | 1% | 2% | 7 |
| 4 | PROTEOMICS | 4427 | 3% | 0% | 28 |
| 5 | CLINICAL PROTEOMICS | 3942 | 0% | 3% | 4 |
| 6 | ACS CHEMICAL BIOLOGY | 3387 | 2% | 1% | 15 |
| 7 | JOURNAL OF PROTEOME RESEARCH | 3303 | 3% | 0% | 25 |
| 8 | EXPERT REVIEW OF PROTEOMICS | 2833 | 1% | 1% | 8 |
| 9 | MOLECULAR CELL | 2127 | 2% | 0% | 19 |
| 10 | NATURE STRUCTURAL & MOLECULAR BIOLOGY | 1805 | 1% | 1% | 11 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SET8 | 379185 | 2% | 73% | 16 | Search SET8 | Search SET8 |
| 2 | HISTONE | 243551 | 14% | 6% | 128 | Search HISTONE | Search HISTONE |
| 3 | PR SET7 | 232758 | 1% | 71% | 10 | Search PR+SET7 | Search PR+SET7 |
| 4 | SPINDLIN1 | 177415 | 1% | 78% | 7 | Search SPINDLIN1 | Search SPINDLIN1 |
| 5 | HISTONE CODE | 133464 | 3% | 16% | 26 | Search HISTONE+CODE | Search HISTONE+CODE |
| 6 | SETD8 | 130344 | 1% | 67% | 6 | Search SETD8 | Search SETD8 |
| 7 | MBT DOMAIN | 104277 | 0% | 80% | 4 | Search MBT+DOMAIN | Search MBT+DOMAIN |
| 8 | HISTONE POST TRANSLATIONAL MODIFICATIONS HPTMS | 97761 | 0% | 100% | 3 | Search HISTONE+POST+TRANSLATIONAL+MODIFICATIONS+HPTMS | Search HISTONE+POST+TRANSLATIONAL+MODIFICATIONS+HPTMS |
| 9 | LYSINE BUTYRYLATION | 97761 | 0% | 100% | 3 | Search LYSINE+BUTYRYLATION | Search LYSINE+BUTYRYLATION |
| 10 | H4K20ME1 | 90515 | 1% | 56% | 5 | Search H4K20ME1 | Search H4K20ME1 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | BRITTON, LMP , GONZALES-COPE, M , ZEE, BM , GARCIA, BA , (2011) BREAKING THE HISTONE CODE WITH QUANTITATIVE MASS SPECTROMETRY.EXPERT REVIEW OF PROTEOMICS. VOL. 8. ISSUE 5. P. 631 -643 | 63 | 58% | 50 |
| 2 | NOBERINI, R , SIGISMONDO, G , BONALDI, T , (2016) THE CONTRIBUTION OF MASS SPECTROMETRY-BASED PROTEOMICS TO UNDERSTANDING EPIGENETICS.EPIGENOMICS. VOL. 8. ISSUE 3. P. 429 -445 | 65 | 46% | 3 |
| 3 | ZHENG, YP , HUANG, XX , KELLEHER, NL , (2016) EPIPROTEOMICS: QUANTITATIVE ANALYSIS OF HISTONE MARKS AND CODES BY MASS SPECTROMETRY.CURRENT OPINION IN CHEMICAL BIOLOGY. VOL. 33. ISSUE . P. 142 -150 | 39 | 85% | 1 |
| 4 | SU, ZL , DENU, JM , (2016) READING THE COMBINATORIAL HISTONE LANGUAGE.ACS CHEMICAL BIOLOGY. VOL. 11. ISSUE 3. P. 564 -574 | 52 | 50% | 4 |
| 5 | VAN NULAND, R , GOZANI, O , (2016) HISTONE H4 LYSINE 20 (H4K20) METHYLATION, EXPANDING THE SIGNALING POTENTIAL OF THE PROTEOME ONE METHYL MOIETY AT A TIME.MOLECULAR & CELLULAR PROTEOMICS. VOL. 15. ISSUE 3. P. 755 -764 | 47 | 54% | 3 |
| 6 | ZHAO, YM , HUANG, H , LIN, S , GARCIA, BA , (2015) QUANTITATIVE PROTEOMIC ANALYSIS OF HISTONE MODIFICATIONS.CHEMICAL REVIEWS. VOL. 115. ISSUE 6. P. 2376 -2418 | 101 | 18% | 38 |
| 7 | SOLDI, M , BREMANG, M , BONALDI, T , (2014) BIOCHEMICAL SYSTEMS APPROACHES FOR THE ANALYSIS OF HISTONE MODIFICATION READOUT.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1839. ISSUE 8. P. 657 -668 | 62 | 46% | 2 |
| 8 | MILOSEVICH, N , HOF, F , (2016) CHEMICAL INHIBITORS OF EPIGENETIC METHYLLYSINE READER PROTEINS.BIOCHEMISTRY. VOL. 55. ISSUE 11. P. 1570 -1583 | 51 | 39% | 4 |
| 9 | MILITE, C , FEOLI, A , VIVIANO, M , RESCIGNO, D , CIANCIULLI, A , BALZANO, AL , MAI, A , CASTELLANO, S , SBARDELLA, G , (2016) THE EMERGING ROLE OF LYSINE METHYLTRANSFERASE SETD8 IN HUMAN DISEASES.CLINICAL EPIGENETICS. VOL. 8. ISSUE . P. - | 48 | 48% | 0 |
| 10 | MILOSEVICH, N , WARMERDAM, Z , HOF, F , (2016) STRUCTURAL ASPECTS OF SMALL-MOLECULE INHIBITION OF METHYLLYSINE READER PROTEINS.FUTURE MEDICINAL CHEMISTRY. VOL. 8. ISSUE 13. P. 1681 -1702 | 50 | 44% | 0 |
Classes with closest relation at Level 1 |
| Rank | Class id | link |
|---|---|---|
| 1 | 14125 | USP22//SET1//UBIQUITIN SPECIFIC PROTEASE 22 |
| 2 | 5481 | ARGININE METHYLATION//PROTEIN ARGININE METHYLTRANSFERASE//PRMT |
| 3 | 8082 | HISTONE DEMETHYLASE//LSD1//HISTONE DEMETHYLATION |
| 4 | 5093 | HISTONE CHAPERONE//HISTONE VARIANTS//ASF1 |
| 5 | 28689 | UHRF1//ICBP90//UHRF2 |
| 6 | 8607 | HISTONE ACETYLTRANSFERASE//GCN5//MOZ |
| 7 | 16660 | COFFIN LOWRY SYNDROME//RSK2//PHOSPHOHISTONE H3 |
| 8 | 13694 | BROMODOMAIN//BRD4//NUT MIDLINE CARCINOMA |
| 9 | 21151 | ING1//ING4//P33ING1 |
| 10 | 1710 | EZH2//POLYCOMB//BMI 1 |