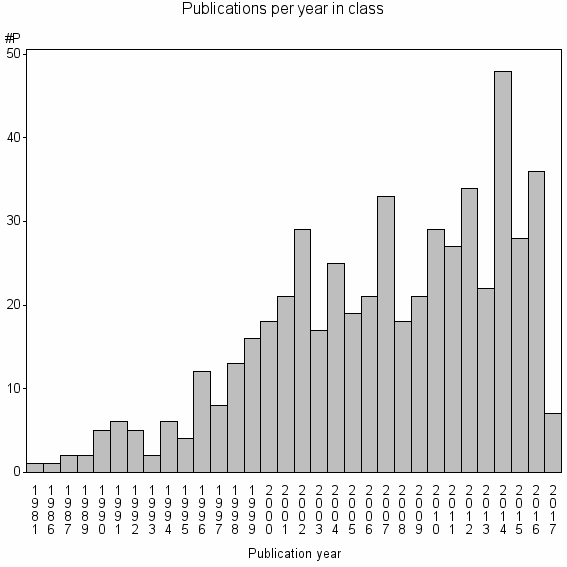
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 18779 | 536 | 42.5 | 87% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 1297 | 2 | RIBOSOME//TRANSLATION TERMINATION//TMRNA | 8599 |
| 18779 | 1 | TMRNA//TRANS TRANSLATION//SMPB | 536 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | TMRNA | authKW | 4312489 | 17% | 84% | 90 |
| 2 | TRANS TRANSLATION | authKW | 3937610 | 13% | 96% | 72 |
| 3 | SMPB | authKW | 1958601 | 7% | 90% | 38 |
| 4 | PEPTIDYL TRNA HYDROLASE | authKW | 1242258 | 5% | 84% | 26 |
| 5 | 10SA RNA | authKW | 1198799 | 4% | 96% | 22 |
| 6 | ABCE1 | authKW | 802495 | 3% | 78% | 18 |
| 7 | SSRA | authKW | 633208 | 3% | 65% | 17 |
| 8 | PELOTA | authKW | 405103 | 1% | 89% | 8 |
| 9 | ARFA | authKW | 256352 | 1% | 75% | 6 |
| 10 | UP JE 2311 | address | 227867 | 1% | 67% | 6 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 4519 | 74% | 0% | 395 |
| 2 | Cell Biology | 952 | 23% | 0% | 122 |
| 3 | Microbiology | 879 | 18% | 0% | 96 |
| 4 | Biophysics | 313 | 10% | 0% | 52 |
| 5 | Genetics & Heredity | 94 | 7% | 0% | 37 |
| 6 | Biochemical Research Methods | 48 | 4% | 0% | 22 |
| 7 | Developmental Biology | 11 | 1% | 0% | 7 |
| 8 | Crystallography | 10 | 2% | 0% | 10 |
| 9 | Andrology | 3 | 0% | 0% | 1 |
| 10 | Biology | 1 | 1% | 0% | 6 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | UP JE 2311 | 227867 | 1% | 67% | 6 |
| 2 | UP JE2311 | 182295 | 1% | 80% | 4 |
| 3 | UP JEUNE EQUIPE 2311IFR 97 | 170903 | 1% | 100% | 3 |
| 4 | BIOCHIM PHARMACEUT | 151904 | 1% | 33% | 8 |
| 5 | UP JEUNE EQUIPE 2311 | 128176 | 1% | 75% | 3 |
| 6 | EQUIPE STRUCT DYNAM MACROMOL | 113936 | 0% | 100% | 2 |
| 7 | ALTHOUSE 401 | 75956 | 0% | 67% | 2 |
| 8 | ANTENNE CHIM SUBST NAT | 75956 | 0% | 67% | 2 |
| 9 | ADV PHOTON GENE | 56968 | 0% | 100% | 1 |
| 10 | BECKMAN B343 | 56968 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA | 7257 | 3% | 1% | 18 |
| 2 | RNA-A PUBLICATION OF THE RNA SOCIETY | 5402 | 2% | 1% | 11 |
| 3 | NUCLEIC ACIDS RESEARCH | 2748 | 8% | 0% | 44 |
| 4 | MOLECULAR MICROBIOLOGY | 2293 | 4% | 0% | 22 |
| 5 | BIOCHIMIE | 1845 | 3% | 0% | 14 |
| 6 | JOURNAL OF MOLECULAR BIOLOGY | 1620 | 5% | 0% | 26 |
| 7 | EMBO JOURNAL | 1442 | 4% | 0% | 21 |
| 8 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 1391 | 1% | 1% | 3 |
| 9 | JOURNAL OF BACTERIOLOGY | 1310 | 5% | 0% | 28 |
| 10 | GENES TO CELLS | 1062 | 1% | 0% | 6 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TMRNA | 4312489 | 17% | 84% | 90 | Search TMRNA | Search TMRNA |
| 2 | TRANS TRANSLATION | 3937610 | 13% | 96% | 72 | Search TRANS+TRANSLATION | Search TRANS+TRANSLATION |
| 3 | SMPB | 1958601 | 7% | 90% | 38 | Search SMPB | Search SMPB |
| 4 | PEPTIDYL TRNA HYDROLASE | 1242258 | 5% | 84% | 26 | Search PEPTIDYL+TRNA+HYDROLASE | Search PEPTIDYL+TRNA+HYDROLASE |
| 5 | 10SA RNA | 1198799 | 4% | 96% | 22 | Search 10SA+RNA | Search 10SA+RNA |
| 6 | ABCE1 | 802495 | 3% | 78% | 18 | Search ABCE1 | Search ABCE1 |
| 7 | SSRA | 633208 | 3% | 65% | 17 | Search SSRA | Search SSRA |
| 8 | PELOTA | 405103 | 1% | 89% | 8 | Search PELOTA | Search PELOTA |
| 9 | ARFA | 256352 | 1% | 75% | 6 | Search ARFA | Search ARFA |
| 10 | RIBOSOME RESCUE | 203454 | 1% | 71% | 5 | Search RIBOSOME+RESCUE | Search RIBOSOME+RESCUE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | HIMENO, H , KURITA, D , MUTO, A , (2014) TMRNA-MEDIATED TRANS-TRANSLATION AS THE MAJOR RIBOSOME RESCUE SYSTEM IN A BACTERIAL CELL.FRONTIERS IN GENETICS. VOL. 5. ISSUE . P. - | 117 | 80% | 10 |
| 2 | HIMENO, H , NAMEKI, N , KURITA, D , MUTO, A , ABO, T , (2015) RIBOSOME RESCUE SYSTEMS IN BACTERIA.BIOCHIMIE. VOL. 114. ISSUE . P. 102 -112 | 109 | 75% | 10 |
| 3 | HIMENO, H , KURITA, D , MUTO, A , (2014) MECHANISM OF TRANS-TRANSLATION REVEALED BY IN VITRO STUDIES.FRONTIERS IN MICROBIOLOGY. VOL. 5. ISSUE . P. - | 92 | 87% | 0 |
| 4 | MOORE, SD , SAUER, RT , (2007) THE TMRNA SYSTEM FOR TRANSLATIONAL SURVEILLANCE AND RIBOSOME RESCUE.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 76. ISSUE . P. 101 -124 | 92 | 70% | 165 |
| 5 | KEILER, KC , (2015) MECHANISMS OF RIBOSOME RESCUE IN BACTERIA.NATURE REVIEWS MICROBIOLOGY. VOL. 13. ISSUE 5. P. 285 -297 | 80 | 62% | 21 |
| 6 | JANSSEN, BD , HAYES, CS , (2012) THE TMRNA RIBOSOME-RESCUE SYSTEM.ADVANCES IN PROTEIN CHEMISTRY AND STRUCTURAL BIOLOGY, VOL 86. VOL. 86. ISSUE . P. 151 -191 | 109 | 55% | 26 |
| 7 | KEILER, KC , (2008) BIOLOGY OF TRANS-TRANSLATION.ANNUAL REVIEW OF MICROBIOLOGY. VOL. 62. ISSUE . P. 133 -151 | 79 | 73% | 102 |
| 8 | SIMMS, CL , THOMAS, EN , ZAHER, HS , (2017) RIBOSOME-BASED QUALITY CONTROL OF MRNA AND NASCENT PEPTIDES.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 8. ISSUE 1. P. - | 68 | 49% | 1 |
| 9 | KEILER, KC , FEAGA, HA , (2014) RESOLVING NONSTOP TRANSLATION COMPLEXES IS A MATTER OF LIFE OR DEATH.JOURNAL OF BACTERIOLOGY. VOL. 196. ISSUE 12. P. 2123 -2130 | 64 | 74% | 7 |
| 10 | DULEBOHN, D , CHOY, J , SUNDERMEIER, T , OKAN, N , KARZAI, AW , (2007) TRANS-TRANSLATION: THE TMRNA-MEDIATED SURVEILLANCE MECHANISM FOR RIBOSOME RESCUE, DIRECTED PROTEIN DEGRADATION, AND NONSTOP MRNA DECAY.BIOCHEMISTRY. VOL. 46. ISSUE 16. P. 4681 -4693 | 72 | 75% | 44 |
Classes with closest relation at Level 1 |