Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
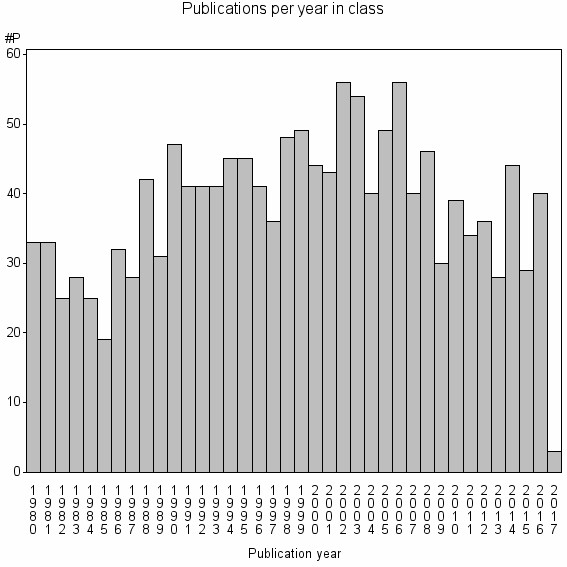
| 6806 | 1441 | 42.4 | 74% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 1297 | 2 | RIBOSOME//TRANSLATION TERMINATION//TMRNA | 8599 |
| 6806 | 1 | TRANSLATION TERMINATION//RELEASE FACTOR//FRAMESHIFT | 1441 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | TRANSLATION TERMINATION | authKW | 1191516 | 7% | 55% | 102 |
| 2 | RELEASE FACTOR | authKW | 622719 | 4% | 57% | 52 |
| 3 | FRAMESHIFT | authKW | 616720 | 6% | 33% | 87 |
| 4 | ERF1 | authKW | 538422 | 2% | 71% | 36 |
| 5 | RIBOSOMAL FRAMESHIFTING | authKW | 317801 | 2% | 50% | 30 |
| 6 | STOP CODON RECOGNITION | authKW | 285485 | 1% | 84% | 16 |
| 7 | RECODING | authKW | 278132 | 2% | 41% | 32 |
| 8 | ERF3 | authKW | 265297 | 2% | 52% | 24 |
| 9 | POLYPEPTIDE RELEASE FACTORS | authKW | 254265 | 1% | 100% | 12 |
| 10 | RNA PSEUDOKNOT | authKW | 250110 | 2% | 54% | 22 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 10969 | 70% | 0% | 1013 |
| 2 | Genetics & Heredity | 1964 | 16% | 0% | 236 |
| 3 | Cell Biology | 1208 | 16% | 0% | 236 |
| 4 | Microbiology | 525 | 9% | 0% | 134 |
| 5 | Virology | 506 | 5% | 0% | 72 |
| 6 | Biophysics | 205 | 5% | 0% | 78 |
| 7 | Biology | 81 | 3% | 0% | 41 |
| 8 | Mycology | 58 | 1% | 0% | 14 |
| 9 | Biotechnology & Applied Microbiology | 31 | 4% | 0% | 51 |
| 10 | Mathematical & Computational Biology | 18 | 1% | 0% | 14 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENET MOL TRADUCT | 67802 | 0% | 80% | 4 |
| 2 | UMR 7098 | 38131 | 0% | 30% | 6 |
| 3 | MAGNET TOMOG SPECT | 28829 | 0% | 19% | 7 |
| 4 | HADASSAH MED VIROL | 28250 | 0% | 67% | 2 |
| 5 | MOL BIOLMINATO KU | 28250 | 0% | 67% | 2 |
| 6 | UPR9073 CNRS | 28250 | 0% | 67% | 2 |
| 7 | UPR41 | 28246 | 0% | 33% | 4 |
| 8 | 513 PARNASSUS AVE | 21189 | 0% | 100% | 1 |
| 9 | AIDS 471 GAKUEN | 21189 | 0% | 100% | 1 |
| 10 | ALLGEMEINE ZOOL GENIT | 21189 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA | 20004 | 3% | 2% | 49 |
| 2 | NUCLEIC ACIDS RESEARCH | 13456 | 11% | 0% | 159 |
| 3 | JOURNAL OF MOLECULAR BIOLOGY | 10708 | 8% | 0% | 109 |
| 4 | RNA-A PUBLICATION OF THE RNA SOCIETY | 9555 | 2% | 2% | 24 |
| 5 | CURRENT GENETICS | 5852 | 2% | 1% | 32 |
| 6 | MOLECULAR BIOLOGY | 3497 | 2% | 1% | 28 |
| 7 | MOLECULAR & GENERAL GENETICS | 3119 | 1% | 1% | 19 |
| 8 | BIOCHIMIE | 2351 | 2% | 0% | 26 |
| 9 | JOURNAL OF BACTERIOLOGY | 2138 | 4% | 0% | 59 |
| 10 | EMBO JOURNAL | 2124 | 3% | 0% | 42 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TRANSLATION TERMINATION | 1191516 | 7% | 55% | 102 | Search TRANSLATION+TERMINATION | Search TRANSLATION+TERMINATION |
| 2 | RELEASE FACTOR | 622719 | 4% | 57% | 52 | Search RELEASE+FACTOR | Search RELEASE+FACTOR |
| 3 | FRAMESHIFT | 616720 | 6% | 33% | 87 | Search FRAMESHIFT | Search FRAMESHIFT |
| 4 | ERF1 | 538422 | 2% | 71% | 36 | Search ERF1 | Search ERF1 |
| 5 | RIBOSOMAL FRAMESHIFTING | 317801 | 2% | 50% | 30 | Search RIBOSOMAL+FRAMESHIFTING | Search RIBOSOMAL+FRAMESHIFTING |
| 6 | STOP CODON RECOGNITION | 285485 | 1% | 84% | 16 | Search STOP+CODON+RECOGNITION | Search STOP+CODON+RECOGNITION |
| 7 | RECODING | 278132 | 2% | 41% | 32 | Search RECODING | Search RECODING |
| 8 | ERF3 | 265297 | 2% | 52% | 24 | Search ERF3 | Search ERF3 |
| 9 | POLYPEPTIDE RELEASE FACTORS | 254265 | 1% | 100% | 12 | Search POLYPEPTIDE+RELEASE+FACTORS | Search POLYPEPTIDE+RELEASE+FACTORS |
| 10 | RNA PSEUDOKNOT | 250110 | 2% | 54% | 22 | Search RNA+PSEUDOKNOT | Search RNA+PSEUDOKNOT |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ATKINS, JF , LOUGHRAN, G , BHATT, PR , FIRTH, AE , BARANOV, PV , (2016) RIBOSOMAL FRAMESHIFTING AND TRANSCRIPTIONAL SLIPPAGE: FROM GENETIC STEGANOGRAPHY AND CRYPTOGRAPHY TO ADVENTITIOUS USE.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE 15. P. 7007 -7078 | 275 | 37% | 1 |
| 2 | HUANG, XL , CHENG, Q , DU, ZH , (2013) A GENOME-WIDE ANALYSIS OF RNA PSEUDOKNOTS THAT STIMULATE EFFICIENT-1 RIBOSOMAL FRAMESHIFTING OR READTHROUGH IN ANIMAL VIRUSES.BIOMED RESEARCH INTERNATIONAL. VOL. . ISSUE . P. - | 59 | 88% | 0 |
| 3 | INGE-VECHTOMOV, S , ZHOURAVLEVA, G , PHILIPPE, M , (2003) EUKARYOTIC RELEASE FACTORS (ERFS) HISTORY.BIOLOGY OF THE CELL. VOL. 95. ISSUE 3-4. P. 195-209 | 84 | 68% | 77 |
| 4 | ATKINS, JF , BJORK, GR , (2009) A GRIPPING TALE OF RIBOSOMAL FRAMESHIFTING: EXTRAGENIC SUPPRESSORS OF FRAMESHIFT MUTATIONS SPOTLIGHT P-SITE REALIGNMENT.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 73. ISSUE 1. P. 178-+ | 114 | 40% | 53 |
| 5 | MOUZAKIS, KD , LANG, AL , VANDER MEULEN, KA , EASTERDAY, PD , BUTCHER, SE , (2013) HIV-1 FRAMESHIFT EFFICIENCY IS PRIMARILY DETERMINED BY THE STABILITY OF BASE PAIRS POSITIONED AT THE MRNA ENTRANCE CHANNEL OF THE RIBOSOME.NUCLEIC ACIDS RESEARCH. VOL. 41. ISSUE 3. P. 1901 -1913 | 57 | 71% | 20 |
| 6 | BARANOV, PV , ATKINS, JF , YORDANOVA, MM , (2015) AUGMENTED GENETIC DECODING: GLOBAL, LOCAL AND TEMPORAL ALTERATIONS OF DECODING PROCESSES AND CODON MEANING.NATURE REVIEWS GENETICS. VOL. 16. ISSUE 9. P. 517 -529 | 82 | 47% | 5 |
| 7 | DINMAN, JD , (2012) MECHANISMS AND IMPLICATIONS OF PROGRAMMED TRANSLATIONAL FRAMESHIFTING.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 3. ISSUE 5. P. 661 -673 | 56 | 65% | 46 |
| 8 | KLAHOLZ, BP , (2011) MOLECULAR RECOGNITION AND CATALYSIS IN TRANSLATION TERMINATION COMPLEXES.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 36. ISSUE 5. P. 282-292 | 57 | 73% | 20 |
| 9 | RODNINA, MV , PESKE, F , CALISKAN, N , (2015) CHANGED IN TRANSLATION: MRNA RECODING BY-1 PROGRAMMED RIBOSOMAL FRAMESHIFTING.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 40. ISSUE 5. P. 265 -274 | 50 | 64% | 14 |
| 10 | DABROWSKI, M , BUKOWY-BIERYLLO, Z , ZIETKIEWICZ, E , (2015) TRANSLATIONAL READTHROUGH POTENTIAL OF NATURAL TERMINATION CODONS IN EUCARYOTES - THE IMPACT OF RNA SEQUENCE.RNA BIOLOGY. VOL. 12. ISSUE 9. P. 950 -958 | 61 | 57% | 5 |
Classes with closest relation at Level 1 |