Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
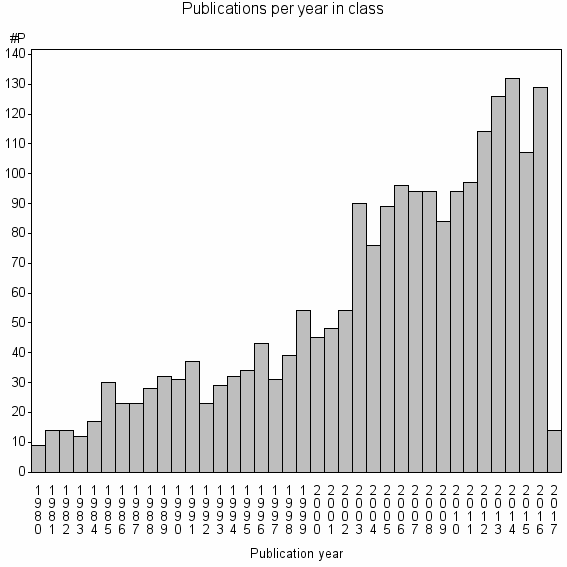
| 2829 | 2138 | 40.6 | 83% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 2829 | 1 | CODON USAGE//CODON USAGE BIAS//SYNONYMOUS CODON USAGE | 2138 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | CODON USAGE | authKW | 1100847 | 10% | 35% | 219 |
| 2 | CODON USAGE BIAS | authKW | 1060078 | 5% | 66% | 113 |
| 3 | SYNONYMOUS CODON USAGE | authKW | 704219 | 3% | 82% | 60 |
| 4 | TRANSLATIONAL SELECTION | authKW | 651628 | 2% | 89% | 51 |
| 5 | CODON BIAS | authKW | 427624 | 4% | 36% | 83 |
| 6 | OPTIMAL CODONS | authKW | 329019 | 1% | 96% | 24 |
| 7 | MUTATIONAL BIAS | authKW | 292893 | 2% | 51% | 40 |
| 8 | AMINO ACID USAGE | authKW | 249245 | 1% | 73% | 24 |
| 9 | MUTATIONAL PRESSURE | authKW | 242210 | 1% | 81% | 21 |
| 10 | EFFECTIVE NUMBER OF CODONS | authKW | 224134 | 1% | 83% | 19 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 15511 | 36% | 0% | 763 |
| 2 | Evolutionary Biology | 11446 | 16% | 0% | 332 |
| 3 | Biochemistry & Molecular Biology | 7354 | 49% | 0% | 1049 |
| 4 | Mathematical & Computational Biology | 6012 | 10% | 0% | 213 |
| 5 | Biotechnology & Applied Microbiology | 1857 | 14% | 0% | 292 |
| 6 | Biology | 1057 | 7% | 0% | 148 |
| 7 | Virology | 754 | 5% | 0% | 107 |
| 8 | Biochemical Research Methods | 701 | 7% | 0% | 148 |
| 9 | Biophysics | 546 | 7% | 0% | 146 |
| 10 | Microbiology | 263 | 6% | 0% | 130 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ORG EVOLUC GENOMA | 215279 | 1% | 54% | 28 |
| 2 | STEM CELLS GENE DRUGS GANSU PROV | 42841 | 0% | 100% | 3 |
| 3 | DISTRIBUTED INFORMAT | 30414 | 0% | 30% | 7 |
| 4 | GUANGZHOU EAST CAMPUS | 28561 | 0% | 100% | 2 |
| 5 | INSECT OURCE ENGN LIAONING PROV | 28561 | 0% | 100% | 2 |
| 6 | PHYSICOTECH SCI TRAINING | 28561 | 0% | 100% | 2 |
| 7 | UMI CNRS UCHILE 2071 | 28561 | 0% | 100% | 2 |
| 8 | CHEMINFORMAT BIOINFORMAT | 21418 | 0% | 50% | 3 |
| 9 | ESCUELA UNIV TECNOL MED | 21418 | 0% | 50% | 3 |
| 10 | O GENET MEJORA ANIM | 21418 | 0% | 50% | 3 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF MOLECULAR EVOLUTION | 71302 | 6% | 4% | 138 |
| 2 | MOLECULAR BIOLOGY AND EVOLUTION | 23015 | 4% | 2% | 95 |
| 3 | DNA RESEARCH | 10825 | 1% | 3% | 22 |
| 4 | NUCLEIC ACIDS RESEARCH | 8182 | 7% | 0% | 152 |
| 5 | JOURNAL OF THEORETICAL BIOLOGY | 6198 | 3% | 1% | 69 |
| 6 | GENOME BIOLOGY AND EVOLUTION | 5559 | 1% | 2% | 23 |
| 7 | GENE | 5465 | 4% | 0% | 87 |
| 8 | EVOLUTIONARY BIOINFORMATICS | 3655 | 0% | 3% | 9 |
| 9 | JOURNAL OF BIOMOLECULAR STRUCTURE & DYNAMICS | 3525 | 1% | 1% | 28 |
| 10 | BMC GENOMICS | 3221 | 2% | 0% | 47 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CODON USAGE | 1100847 | 10% | 35% | 219 | Search CODON+USAGE | Search CODON+USAGE |
| 2 | CODON USAGE BIAS | 1060078 | 5% | 66% | 113 | Search CODON+USAGE+BIAS | Search CODON+USAGE+BIAS |
| 3 | SYNONYMOUS CODON USAGE | 704219 | 3% | 82% | 60 | Search SYNONYMOUS+CODON+USAGE | Search SYNONYMOUS+CODON+USAGE |
| 4 | TRANSLATIONAL SELECTION | 651628 | 2% | 89% | 51 | Search TRANSLATIONAL+SELECTION | Search TRANSLATIONAL+SELECTION |
| 5 | CODON BIAS | 427624 | 4% | 36% | 83 | Search CODON+BIAS | Search CODON+BIAS |
| 6 | OPTIMAL CODONS | 329019 | 1% | 96% | 24 | Search OPTIMAL+CODONS | Search OPTIMAL+CODONS |
| 7 | MUTATIONAL BIAS | 292893 | 2% | 51% | 40 | Search MUTATIONAL+BIAS | Search MUTATIONAL+BIAS |
| 8 | AMINO ACID USAGE | 249245 | 1% | 73% | 24 | Search AMINO+ACID+USAGE | Search AMINO+ACID+USAGE |
| 9 | MUTATIONAL PRESSURE | 242210 | 1% | 81% | 21 | Search MUTATIONAL+PRESSURE | Search MUTATIONAL+PRESSURE |
| 10 | EFFECTIVE NUMBER OF CODONS | 224134 | 1% | 83% | 19 | Search EFFECTIVE+NUMBER+OF+CODONS | Search EFFECTIVE+NUMBER+OF+CODONS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | PLOTKIN, JB , KUDLA, G , (2011) SYNONYMOUS BUT NOT THE SAME: THE CAUSES AND CONSEQUENCES OF CODON BIAS.NATURE REVIEWS GENETICS. VOL. 12. ISSUE 1. P. 32-42 | 83 | 63% | 365 |
| 2 | QUAX, TEF , CLAASSENS, NJ , SOLL, D , VAN DER OOST, J , (2015) CODON BIAS AS A MEANS TO FINE-TUNE GENE EXPRESSION.MOLECULAR CELL. VOL. 59. ISSUE 2. P. 149 -161 | 61 | 76% | 30 |
| 3 | SUPEK, F , (2016) THE CODE OF SILENCE: WIDESPREAD ASSOCIATIONS BETWEEN SYNONYMOUS CODON BIASES AND GENE FUNCTION.JOURNAL OF MOLECULAR EVOLUTION. VOL. 82. ISSUE 1. P. 65 -73 | 71 | 71% | 4 |
| 4 | KOMAR, AA , (2016) THE YIN AND YANG OF CODON USAGE.HUMAN MOLECULAR GENETICS. VOL. 25. ISSUE . P. R77 -R85 | 73 | 74% | 2 |
| 5 | BEHURA, SK , SEVERSON, DW , (2013) CODON USAGE BIAS: CAUSATIVE FACTORS, QUANTIFICATION METHODS AND GENOME-WIDE PATTERNS: WITH EMPHASIS ON INSECT GENOMES.BIOLOGICAL REVIEWS. VOL. 88. ISSUE 1. P. 49 -61 | 70 | 69% | 35 |
| 6 | CHANEY, JL , CLARK, PL , (2015) ROLES FOR SYNONYMOUS CODON USAGE IN PROTEIN BIOGENESIS.ANNUAL REVIEW OF BIOPHYSICS, VOL 44. VOL. 44. ISSUE . P. 143 -166 | 75 | 54% | 18 |
| 7 | GINGOLD, H , PILPEL, Y , (2011) DETERMINANTS OF TRANSLATION EFFICIENCY AND ACCURACY.MOLECULAR SYSTEMS BIOLOGY. VOL. 7. ISSUE . P. - | 68 | 55% | 134 |
| 8 | ROY, A , MUKHOPADHYAY, S , SARKAR, I , SEN, A , (2015) COMPARATIVE INVESTIGATION OF THE VARIOUS DETERMINANTS THAT INFLUENCE THE CODON AND AMINO ACID USAGE PATTERNS IN THE GENUS BIFIDOBACTERIUM.WORLD JOURNAL OF MICROBIOLOGY & BIOTECHNOLOGY. VOL. 31. ISSUE 6. P. 959 -981 | 66 | 72% | 1 |
| 9 | ANGOV, E , (2011) CODON USAGE: NATURE'S ROADMAP TO EXPRESSION AND FOLDING OF PROTEINS.BIOTECHNOLOGY JOURNAL. VOL. 6. ISSUE 6. P. 650 -659 | 63 | 65% | 58 |
| 10 | ZUR, H , TULLER, T , (2016) PREDICTIVE BIOPHYSICAL MODELING AND UNDERSTANDING OF THE DYNAMICS OF MRNA TRANSLATION AND ITS EVOLUTION.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE 19. P. 9031 -9049 | 91 | 48% | 0 |
Classes with closest relation at Level 1 |