Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
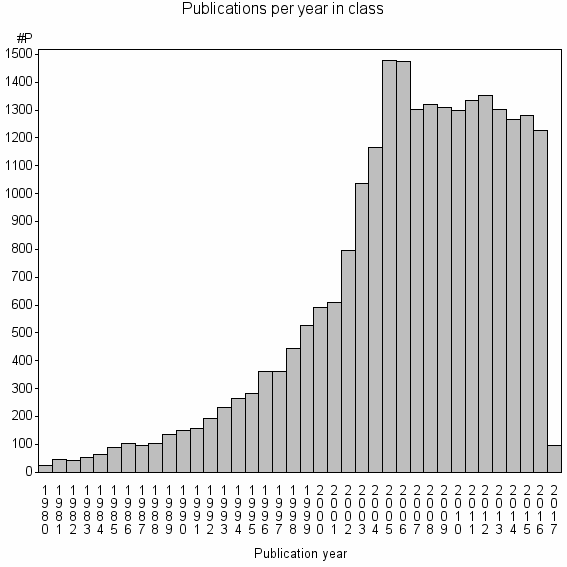
| 126 | 23963 | 39.8 | 78% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | MOLECULAR BIOLOGY AND EVOLUTION | journal | 300029 | 5% | 21% | 1148 |
| 2 | BIOINFORMATICS | journal | 289584 | 7% | 15% | 1560 |
| 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY | WoSSC | 256789 | 19% | 5% | 4585 |
| 4 | STANDARDS IN GENOMIC SCIENCES | journal | 211504 | 1% | 50% | 334 |
| 5 | EVOLUTIONARY BIOLOGY | WoSSC | 162653 | 17% | 3% | 4177 |
| 6 | BMC BIOINFORMATICS | journal | 156804 | 4% | 13% | 965 |
| 7 | GENETICS & HEREDITY | WoSSC | 156517 | 34% | 2% | 8134 |
| 8 | BIOL DATA MANAGEMENT TECHNOL | address | 153974 | 1% | 70% | 174 |
| 9 | SYSTEMATIC BIOLOGY | journal | 152688 | 2% | 30% | 401 |
| 10 | JOURNAL OF COMPUTATIONAL BIOLOGY | journal | 139448 | 2% | 27% | 414 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 256789 | 19% | 5% | 4585 |
| 2 | Evolutionary Biology | 162653 | 17% | 3% | 4177 |
| 3 | Genetics & Heredity | 156517 | 34% | 2% | 8134 |
| 4 | Biotechnology & Applied Microbiology | 70375 | 24% | 1% | 5685 |
| 5 | Biochemical Research Methods | 61129 | 17% | 1% | 4176 |
| 6 | Biochemistry & Molecular Biology | 35407 | 34% | 0% | 8161 |
| 7 | Computer Science, Interdisciplinary Applications | 6568 | 5% | 1% | 1217 |
| 8 | Biology | 6462 | 5% | 1% | 1273 |
| 9 | Statistics & Probability | 3653 | 4% | 0% | 848 |
| 10 | Microbiology | 3109 | 6% | 0% | 1486 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOL DATA MANAGEMENT TECHNOL | 153974 | 1% | 70% | 174 |
| 2 | BIOTECHNOL INFORMAT | 94876 | 2% | 19% | 403 |
| 3 | BIOINFORMAT | 60510 | 3% | 6% | 820 |
| 4 | EVOLUZ MOL | 47999 | 0% | 88% | 43 |
| 5 | LIB MED | 39738 | 1% | 16% | 193 |
| 6 | COMP SCI | 28834 | 8% | 1% | 1945 |
| 7 | ORG EVOLUC GENOMA | 28261 | 0% | 65% | 34 |
| 8 | EMBL OUTSTN | 27787 | 0% | 31% | 71 |
| 9 | EVOLUTIONARY BIOINFORMAT | 21484 | 0% | 65% | 26 |
| 10 | JOINT GENOME | 20341 | 0% | 18% | 90 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR BIOLOGY AND EVOLUTION | 300029 | 5% | 21% | 1148 |
| 2 | BIOINFORMATICS | 289584 | 7% | 15% | 1560 |
| 3 | STANDARDS IN GENOMIC SCIENCES | 211504 | 1% | 50% | 334 |
| 4 | BMC BIOINFORMATICS | 156804 | 4% | 13% | 965 |
| 5 | SYSTEMATIC BIOLOGY | 152688 | 2% | 30% | 401 |
| 6 | JOURNAL OF COMPUTATIONAL BIOLOGY | 139448 | 2% | 27% | 414 |
| 7 | JOURNAL OF MOLECULAR EVOLUTION | 118726 | 2% | 16% | 598 |
| 8 | BIOTECHFORUM ( BTF ) | 100818 | 2% | 18% | 454 |
| 9 | NUCLEIC ACIDS RESEARCH | 98325 | 7% | 5% | 1763 |
| 10 | COMPUTER APPLICATIONS IN THE BIOSCIENCES | 86419 | 1% | 27% | 249 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CODON USAGE | 132707 | 1% | 41% | 255 | Search CODON+USAGE | Search CODON+USAGE |
| 2 | ISOCHORES | 115850 | 1% | 68% | 135 | Search ISOCHORES | Search ISOCHORES |
| 3 | CODON USAGE BIAS | 110027 | 1% | 71% | 122 | Search CODON+USAGE+BIAS | Search CODON+USAGE+BIAS |
| 4 | PHYLOGENETIC NETWORK | 100597 | 0% | 68% | 117 | Search PHYLOGENETIC+NETWORK | Search PHYLOGENETIC+NETWORK |
| 5 | GEBA | 89174 | 0% | 75% | 94 | Search GEBA | Search GEBA |
| 6 | GC CONTENT | 74949 | 1% | 42% | 140 | Search GC+CONTENT | Search GC+CONTENT |
| 7 | MULTIPLE SEQUENCE ALIGNMENT | 71294 | 1% | 40% | 139 | Search MULTIPLE+SEQUENCE+ALIGNMENT | Search MULTIPLE+SEQUENCE+ALIGNMENT |
| 8 | SYNONYMOUS CODON USAGE | 62722 | 0% | 82% | 60 | Search SYNONYMOUS+CODON+USAGE | Search SYNONYMOUS+CODON+USAGE |
| 9 | SEQUENCE ALIGNMENT | 60505 | 1% | 27% | 174 | Search SEQUENCE+ALIGNMENT | Search SEQUENCE+ALIGNMENT |
| 10 | TRANSLATIONAL SELECTION | 60347 | 0% | 91% | 52 | Search TRANSLATIONAL+SELECTION | Search TRANSLATIONAL+SELECTION |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ALTSCHUL, SF , MADDEN, TL , SCHAFFER, AA , ZHANG, JH , ZHANG, Z , MILLER, W , LIPMAN, DJ , (1997) GAPPED BLAST AND PSI-BLAST: A NEW GENERATION OF PROTEIN DATABASE SEARCH PROGRAMS.NUCLEIC ACIDS RESEARCH. VOL. 25. ISSUE 17. P. 3389-3402 | 43 | 70% | 41839 |
| 2 | MORRISON, DA , (2006) PHYLOGENETIC ANALYSES OF PARASITES IN THE NEW MILLENNIUM.ADVANCES IN PARASITOLOGY, VOL 63. VOL. 63. ISSUE . P. 1 -124 | 246 | 73% | 7 |
| 3 | KUMAR, S , FILIPSKI, AJ , BATTISTUZZI, FU , POND, SLK , TAMURA, K , (2012) STATISTICS AND TRUTH IN PHYLOGENOMICS.MOLECULAR BIOLOGY AND EVOLUTION. VOL. 29. ISSUE 2. P. 457 -472 | 127 | 73% | 54 |
| 4 | ROGOZIN, IB , CARMEL, L , CSUROS, M , KOONIN, EV , (2012) ORIGIN AND EVOLUTION OF SPLICEOSOMAL INTRONS.BIOLOGY DIRECT. VOL. 7. ISSUE . P. - | 153 | 56% | 69 |
| 5 | GUINDON, S , GASCUEL, O , (2003) A SIMPLE, FAST, AND ACCURATE ALGORITHM TO ESTIMATE LARGE PHYLOGENIES BY MAXIMUM LIKELIHOOD.SYSTEMATIC BIOLOGY. VOL. 52. ISSUE 5. P. 696-704 | 29 | 83% | 9889 |
| 6 | PLOTKIN, JB , KUDLA, G , (2011) SYNONYMOUS BUT NOT THE SAME: THE CAUSES AND CONSEQUENCES OF CODON BIAS.NATURE REVIEWS GENETICS. VOL. 12. ISSUE 1. P. 32-42 | 85 | 65% | 365 |
| 7 | GUINDON, S , DUFAYARD, JF , LEFORT, V , ANISIMOVA, M , HORDIJK, W , GASCUEL, O , (2010) NEW ALGORITHMS AND METHODS TO ESTIMATE MAXIMUM-LIKELIHOOD PHYLOGENIES: ASSESSING THE PERFORMANCE OF PHYML 3.0.SYSTEMATIC BIOLOGY. VOL. 59. ISSUE 3. P. 307 -321 | 25 | 86% | 3880 |
| 8 | SZOLLOSI, GJ , TANNIER, E , DAUBIN, V , BOUSSAU, B , (2015) THE INFERENCE OF GENE TREES WITH SPECIES TREES.SYSTEMATIC BIOLOGY. VOL. 64. ISSUE 1. P. E42 -E62 | 92 | 77% | 23 |
| 9 | MORRISON, DA , (2006) MULTIPLE SEQUENCE ALIGNMENT FOR PHYLOGENETIC PURPOSES.AUSTRALIAN SYSTEMATIC BOTANY. VOL. 19. ISSUE 6. P. 479 -539 | 184 | 57% | 73 |
| 10 | EDWARDS, SV , XI, ZX , JANKE, A , FAIRCLOTH, BC , MCCORMACK, JE , GLENN, TC , ZHONG, BJ , WU, SY , LEMMON, EM , LEMMON, AR , ET AL (2016) IMPLEMENTING AND TESTING THE MULTISPECIES COALESCENT MODEL: A VALUABLE PARADIGM FOR PHYLOGENOMICS.MOLECULAR PHYLOGENETICS AND EVOLUTION. VOL. 94. ISSUE . P. 447 -462 | 71 | 73% | 12 |
Classes with closest relation at Level 2 |