Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
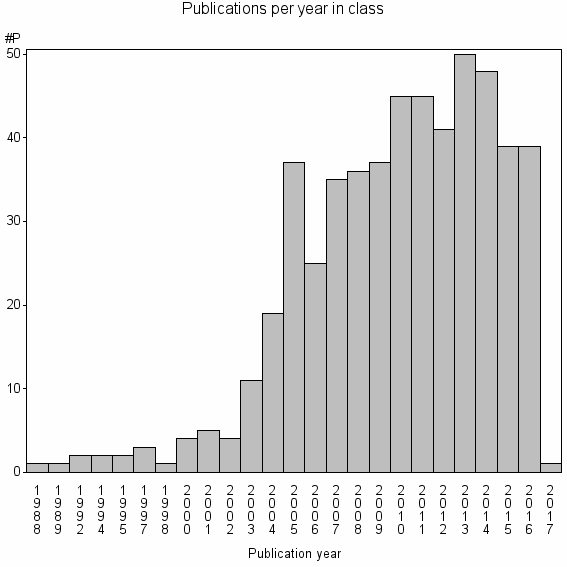
| 18864 | 533 | 26.5 | 71% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 18864 | 1 | DATABASE LIFE SCI//DDBJ//EMTRAIN | 533 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | DATABASE LIFE SCI | address | 144398 | 2% | 23% | 11 |
| 2 | DDBJ | address | 141007 | 2% | 31% | 8 |
| 3 | EMTRAIN | address | 114577 | 0% | 100% | 2 |
| 4 | MIABI | authKW | 114577 | 0% | 100% | 2 |
| 5 | MOUSE GENOM INFORMAT GRP | address | 114577 | 0% | 100% | 2 |
| 6 | PHARMATRAIN | address | 114577 | 0% | 100% | 2 |
| 7 | RD SCI RELAT | address | 114577 | 0% | 100% | 2 |
| 8 | RESOURCEOME | authKW | 114577 | 0% | 100% | 2 |
| 9 | VECTORBASE | address | 114577 | 0% | 100% | 2 |
| 10 | DNA DATA BANK J AN | address | 103447 | 3% | 11% | 17 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 23952 | 39% | 0% | 207 |
| 2 | Biochemical Research Methods | 5182 | 33% | 0% | 176 |
| 3 | Biotechnology & Applied Microbiology | 3162 | 33% | 0% | 176 |
| 4 | Biochemistry & Molecular Biology | 813 | 35% | 0% | 184 |
| 5 | Genetics & Heredity | 131 | 8% | 0% | 42 |
| 6 | Computer Science, Theory & Methods | 104 | 5% | 0% | 24 |
| 7 | Computer Science, Information Systems | 53 | 3% | 0% | 17 |
| 8 | Computer Science, Interdisciplinary Applications | 47 | 3% | 0% | 17 |
| 9 | Medical Informatics | 34 | 1% | 0% | 6 |
| 10 | Computer Science, Software Engineering | 31 | 2% | 0% | 12 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DATABASE LIFE SCI | 144398 | 2% | 23% | 11 |
| 2 | DDBJ | 141007 | 2% | 31% | 8 |
| 3 | EMTRAIN | 114577 | 0% | 100% | 2 |
| 4 | MOUSE GENOM INFORMAT GRP | 114577 | 0% | 100% | 2 |
| 5 | PHARMATRAIN | 114577 | 0% | 100% | 2 |
| 6 | RD SCI RELAT | 114577 | 0% | 100% | 2 |
| 7 | VECTORBASE | 114577 | 0% | 100% | 2 |
| 8 | DNA DATA BANK J AN | 103447 | 3% | 11% | 17 |
| 9 | ENGN ESAT SCD | 76383 | 0% | 67% | 2 |
| 10 | BIOSCI DATABASE | 70503 | 1% | 31% | 4 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DATABASE-THE JOURNAL OF BIOLOGICAL DATABASES AND CURATION | 54952 | 5% | 4% | 25 |
| 2 | JOURNAL OF BIOMEDICAL SEMANTICS | 34066 | 2% | 5% | 12 |
| 3 | NUCLEIC ACIDS RESEARCH | 30253 | 27% | 0% | 144 |
| 4 | BIOINFORMATICS | 28670 | 14% | 1% | 73 |
| 5 | BMC BIOINFORMATICS | 27468 | 11% | 1% | 60 |
| 6 | BRIEFINGS IN BIOINFORMATICS | 23789 | 3% | 2% | 17 |
| 7 | LECTURE NOTES IN BIOINFORMATICS | 2934 | 0% | 3% | 2 |
| 8 | BIOTECHFORUM ( BTF ) | 1073 | 1% | 0% | 7 |
| 9 | MYCOKEYS | 922 | 0% | 2% | 1 |
| 10 | GENOME MEDICINE | 782 | 1% | 0% | 3 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MIABI | 114577 | 0% | 100% | 2 | Search MIABI | Search MIABI |
| 2 | RESOURCEOME | 114577 | 0% | 100% | 2 | Search RESOURCEOME | Search RESOURCEOME |
| 3 | GENOME BROWSER | 82486 | 1% | 24% | 6 | Search GENOME+BROWSER | Search GENOME+BROWSER |
| 4 | BIOHACKATHON | 76383 | 0% | 67% | 2 | Search BIOHACKATHON | Search BIOHACKATHON |
| 5 | BIOMOBY | 76383 | 0% | 67% | 2 | Search BIOMOBY | Search BIOMOBY |
| 6 | SADI | 73653 | 1% | 43% | 3 | Search SADI | Search SADI |
| 7 | 3 3 CHOLAMIDOPROPYLDIMETHYLAMMONIO 1 PROPANE SULFONATE CHAPS | 57288 | 0% | 100% | 1 | Search 3+3+CHOLAMIDOPROPYLDIMETHYLAMMONIO+1+PROPANE+SULFONATE+CHAPS | Search 3+3+CHOLAMIDOPROPYLDIMETHYLAMMONIO+1+PROPANE+SULFONATE+CHAPS |
| 8 | 3D EM DAS | 57288 | 0% | 100% | 1 | Search 3D+EM+DAS | Search 3D+EM+DAS |
| 9 | ADAPTIVE CONFLICT | 57288 | 0% | 100% | 1 | Search ADAPTIVE+CONFLICT | Search ADAPTIVE+CONFLICT |
| 10 | ANALYSIS ARTEFACT | 57288 | 0% | 100% | 1 | Search ANALYSIS+ARTEFACT | Search ANALYSIS+ARTEFACT |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | BENSON, DA , CLARK, K , KARSCH-MIZRACHI, I , LIPMAN, DJ , OSTELL, J , SAYERS, EW , (2015) GENBANK.NUCLEIC ACIDS RESEARCH. VOL. 43. ISSUE D1. P. D30 -D35 | 10 | 71% | 94 |
| 2 | CLARK, K , KARSCH-MIZRACHI, I , LIPMAN, DJ , OSTELL, J , SAYERS, EW , (2016) GENBANK.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE D1. P. D67 -D72 | 9 | 69% | 12 |
| 3 | BENSON, DA , CAVANAUGH, M , CLARK, K , KARSCH-MIZRACHI, I , LIPMAN, DJ , OSTELL, J , SAYERS, EW , (2013) GENBANK.NUCLEIC ACIDS RESEARCH. VOL. 41. ISSUE D1. P. D36-D42 | 6 | 60% | 532 |
| 4 | MASHIMA, J , KODAMA, Y , KOSUGE, T , FUJISAWA, T , KATAYAMA, T , NAGASAKI, H , OKUDA, Y , KAMINUMA, E , OGASAWARA, O , OKUBO, K , ET AL (2016) DNA DATA BANK OF JAPAN (DDBJ) PROGRESS REPORT.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE D1. P. D51 -D57 | 14 | 58% | 2 |
| 5 | KODAMA, Y , MASHIMA, J , KOSUGE, T , KATAYAMA, T , FUJISAWA, T , KAMINUMA, E , OGASAWARA, O , OKUBO, K , TAKAGI, T , NAKAMURA, Y , (2015) THE DDBJ JAPANESE GENOTYPE-PHENOTYPE ARCHIVE FOR GENETIC AND PHENOTYPIC HUMAN DATA.NUCLEIC ACIDS RESEARCH. VOL. 43. ISSUE D1. P. D18 -D22 | 16 | 52% | 5 |
| 6 | KOSUGE, T , MASHIMA, J , KODAMA, Y , FUJISAWA, T , KAMINUMA, E , OGASAWARA, O , OKUBO, K , TAKAGI, T , NAKAMURA, Y , (2014) DDBJ PROGRESS REPORT: A NEW SUBMISSION SYSTEM FOR LEADING TO A CORRECT ANNOTATION.NUCLEIC ACIDS RESEARCH. VOL. 42. ISSUE D1. P. D44-D49 | 13 | 59% | 16 |
| 7 | MORENO, BG , PEYPOCH, XM , (2011) GENEXP: AN INTERACTIVE WEB-BASED GENOMIC DAS CLIENT WITH CLIENT-SIDE DATA RENDERING.PLOS ONE. VOL. 6. ISSUE 7. P. - | 11 | 85% | 1 |
| 8 | BHAGAT, J , TANOH, F , NZUOBONTANE, E , LAURENT, T , ORLOWSKI, J , ROOS, M , WOLSTENCROFT, K , ALEKSEJEVS, S , STEVENS, R , PETTIFER, S , ET AL (2010) BIOCATALOGUE: A UNIVERSAL CATALOGUE OF WEB SERVICES FOR THE LIFE SCIENCES.NUCLEIC ACIDS RESEARCH. VOL. 38. ISSUE . P. W689 -W694 | 11 | 69% | 63 |
| 9 | BENSON, DA , CLARK, K , KARSCH-MIZRACHI, I , LIPMAN, DJ , OSTELL, J , SAYERS, EW , (2014) GENBANK.NUCLEIC ACIDS RESEARCH. VOL. 42. ISSUE D1. P. D32-D37 | 7 | 64% | 120 |
| 10 | MCWILLIAM, H , LI, WZ , ULUDAG, M , SQUIZZATO, S , PARK, YM , BUSO, N , COWLEY, AP , LOPEZ, R , (2013) ANALYSIS TOOL WEB SERVICES FROM THE EMBL-EBI.NUCLEIC ACIDS RESEARCH. VOL. 41. ISSUE W1. P. W597-W600 | 9 | 36% | 350 |
Classes with closest relation at Level 1 |