Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
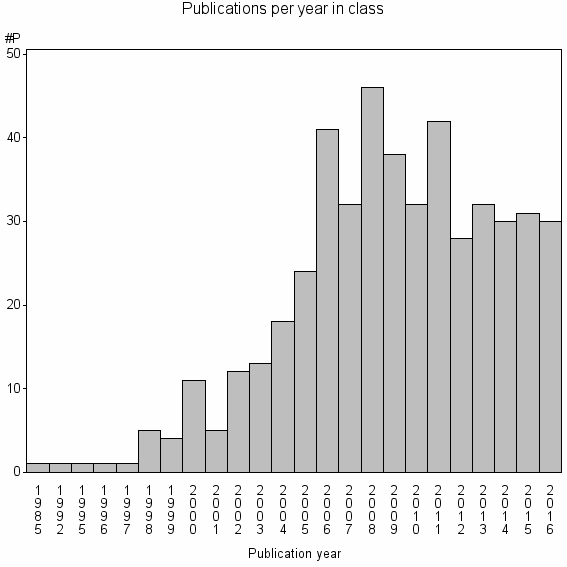
| 20039 | 479 | 34.9 | 81% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 20039 | 1 | ORTHOLOGY//BIDIRECTIONAL BEST HIT//GENOME FUNCTIONAL PREDICTION | 479 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | ORTHOLOGY | authKW | 356100 | 5% | 21% | 26 |
| 2 | BIDIRECTIONAL BEST HIT | authKW | 143430 | 1% | 75% | 3 |
| 3 | GENOME FUNCTIONAL PREDICTION | authKW | 127494 | 0% | 100% | 2 |
| 4 | GERMANY MOL MED PARTNERSHIP UNIT | address | 127494 | 0% | 100% | 2 |
| 5 | GERMANY MOL MED PARTNERSHIP UNIT MMPU | address | 127494 | 0% | 100% | 2 |
| 6 | INPARALOG | authKW | 127494 | 0% | 100% | 2 |
| 7 | RECIPROCAL BEST HIT | authKW | 127494 | 0% | 100% | 2 |
| 8 | UNIREF | authKW | 127494 | 0% | 100% | 2 |
| 9 | XENOLOG | authKW | 127494 | 0% | 100% | 2 |
| 10 | BIOINFORMAT GENOM PROGRAMME | address | 110546 | 3% | 12% | 14 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 23184 | 40% | 0% | 193 |
| 2 | Biochemical Research Methods | 6290 | 38% | 0% | 183 |
| 3 | Biotechnology & Applied Microbiology | 5295 | 44% | 0% | 213 |
| 4 | Genetics & Heredity | 1446 | 24% | 0% | 113 |
| 5 | Evolutionary Biology | 822 | 9% | 0% | 43 |
| 6 | Biochemistry & Molecular Biology | 759 | 35% | 0% | 168 |
| 7 | Mycology | 6 | 1% | 0% | 3 |
| 8 | Computer Science, Theory & Methods | 4 | 1% | 0% | 7 |
| 9 | Computer Science, Interdisciplinary Applications | 2 | 1% | 0% | 6 |
| 10 | Computer Science, Software Engineering | 1 | 1% | 0% | 4 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GERMANY MOL MED PARTNERSHIP UNIT | 127494 | 0% | 100% | 2 |
| 2 | GERMANY MOL MED PARTNERSHIP UNIT MMPU | 127494 | 0% | 100% | 2 |
| 3 | BIOINFORMAT GENOM PROGRAMME | 110546 | 3% | 12% | 14 |
| 4 | BERKELEY PHYLOGENOM GRP | 84995 | 0% | 67% | 2 |
| 5 | ANDALUZ BIOL DESARROLLO CABDCSICJA | 63747 | 0% | 100% | 1 |
| 6 | ASSAYS BIOINFORMAT | 63747 | 0% | 100% | 1 |
| 7 | BGBP UMR 5558 | 63747 | 0% | 100% | 1 |
| 8 | BINFORMAT | 63747 | 0% | 100% | 1 |
| 9 | BIOINFORMAT COMPUTAT GENOMIX BIOBIX | 63747 | 0% | 100% | 1 |
| 10 | BIOL STRUCT GENOM INSERM | 63747 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BMC BIOINFORMATICS | 32655 | 13% | 1% | 62 |
| 2 | BIOINFORMATICS | 30189 | 15% | 1% | 71 |
| 3 | EVOLUTIONARY BIOINFORMATICS | 12936 | 2% | 3% | 8 |
| 4 | NUCLEIC ACIDS RESEARCH | 12516 | 18% | 0% | 88 |
| 5 | BRIEFINGS IN BIOINFORMATICS | 5854 | 2% | 1% | 8 |
| 6 | GENOME BIOLOGY AND EVOLUTION | 3813 | 2% | 1% | 9 |
| 7 | BMC GENOMICS | 3198 | 5% | 0% | 22 |
| 8 | DATABASE-THE JOURNAL OF BIOLOGICAL DATABASES AND CURATION | 2438 | 1% | 1% | 5 |
| 9 | BIOTECHFORUM ( BTF ) | 1982 | 2% | 0% | 9 |
| 10 | PLOS COMPUTATIONAL BIOLOGY | 1967 | 3% | 0% | 12 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | ORTHOLOGY | 356100 | 5% | 21% | 26 | Search ORTHOLOGY | Search ORTHOLOGY |
| 2 | BIDIRECTIONAL BEST HIT | 143430 | 1% | 75% | 3 | Search BIDIRECTIONAL+BEST+HIT | Search BIDIRECTIONAL+BEST+HIT |
| 3 | GENOME FUNCTIONAL PREDICTION | 127494 | 0% | 100% | 2 | Search GENOME+FUNCTIONAL+PREDICTION | Search GENOME+FUNCTIONAL+PREDICTION |
| 4 | INPARALOG | 127494 | 0% | 100% | 2 | Search INPARALOG | Search INPARALOG |
| 5 | RECIPROCAL BEST HIT | 127494 | 0% | 100% | 2 | Search RECIPROCAL+BEST+HIT | Search RECIPROCAL+BEST+HIT |
| 6 | UNIREF | 127494 | 0% | 100% | 2 | Search UNIREF | Search UNIREF |
| 7 | XENOLOG | 127494 | 0% | 100% | 2 | Search XENOLOG | Search XENOLOG |
| 8 | HOMOLOGY INFERENCE | 84995 | 0% | 67% | 2 | Search HOMOLOGY+INFERENCE | Search HOMOLOGY+INFERENCE |
| 9 | ORTHOLOG | 84195 | 3% | 11% | 12 | Search ORTHOLOG | Search ORTHOLOG |
| 10 | ACNUC | 63747 | 0% | 100% | 1 | Search ACNUC | Search ACNUC |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | POWELL, S , SZKLARCZYK, D , TRACHANA, K , ROTH, A , KUHN, M , MULLER, J , ARNOLD, R , RATTEI, T , LETUNIC, I , DOERKS, T , ET AL (2012) EGGNOG V3.0: ORTHOLOGOUS GROUPS COVERING 1133 ORGANISMS AT 41 DIFFERENT TAXONOMIC RANGES.NUCLEIC ACIDS RESEARCH. VOL. 40. ISSUE D1. P. D284 -D289 | 30 | 67% | 146 |
| 2 | HUERTA-CEPAS, J , SZKLARCZYK, D , FORSLUND, K , COOK, H , HELLER, D , WALTER, MC , RATTEI, T , MENDE, DR , SUNAGAWA, S , KUHN, M , ET AL (2016) EGGNOG 4.5: A HIERARCHICAL ORTHOLOGY FRAMEWORK WITH IMPROVED FUNCTIONAL ANNOTATIONS FOR EUKARYOTIC, PROKARYOTIC AND VIRAL SEQUENCES.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE D1. P. D286 -D293 | 23 | 48% | 33 |
| 3 | KRISTENSEN, DM , WOLF, YI , MUSHEGIAN, AR , KOONIN, EV , (2011) COMPUTATIONAL METHODS FOR GENE ORTHOLOGY INFERENCE.BRIEFINGS IN BIOINFORMATICS. VOL. 12. ISSUE 5. P. 379 -391 | 50 | 42% | 60 |
| 4 | ALTENHOFF, AM , BOECKMANN, B , CAPELLA-GUTIERREZ, S , DALQUEN, DA , DELUCA, T , FORSLUND, K , HUERTA-CEPAS, J , LINARD, B , PEREIRA, C , PRYSZCZ, LP , ET AL (2016) STANDARDIZED BENCHMARKING IN THE QUEST FOR ORTHOLOGS.NATURE METHODS. VOL. 13. ISSUE 5. P. 425 -+ | 33 | 61% | 1 |
| 5 | ALTENHOFF, AM , SCHNEIDER, A , GONNET, GH , DESSIMOZ, C , (2011) OMA 2011: ORTHOLOGY INFERENCE AMONG 1000 COMPLETE GENOMES.NUCLEIC ACIDS RESEARCH. VOL. 39. ISSUE . P. D289 -D294 | 20 | 71% | 91 |
| 6 | ALTENHOFF, AM , GIL, M , GONNET, GH , DESSIMOZ, C , (2013) INFERRING HIERARCHICAL ORTHOLOGOUS GROUPS FROM ORTHOLOGOUS GENE PAIRS.PLOS ONE. VOL. 8. ISSUE 1. P. - | 21 | 72% | 24 |
| 7 | WHITESIDE, MD , WINSOR, GL , LAIRD, MR , BRINKMAN, FSL , (2013) ORTHOLUGEDB: A BACTERIAL AND ARCHAEAL ORTHOLOGY RESOURCE FOR IMPROVED COMPARATIVE GENOMIC ANALYSIS.NUCLEIC ACIDS RESEARCH. VOL. 41. ISSUE D1. P. D366-D376 | 26 | 57% | 23 |
| 8 | KUZNIAR, A , VAN HAM, RCHJ , PONGOR, S , LEUNISSEN, JAM , (2008) THE QUEST FOR ORTHOLOGS: FINDING THE CORRESPONDING GENE ACROSS GENOMES.TRENDS IN GENETICS. VOL. 24. ISSUE 11. P. 539 -551 | 31 | 44% | 120 |
| 9 | ALTENHOFF, AM , SKUNCA, N , GLOVER, N , TRAIN, CM , SUEKI, A , PILIZOTA, I , GORI, K , TOMICZEK, B , MULLER, S , REDESTIG, H , ET AL (2015) THE OMA ORTHOLOGY DATABASE IN 2015: FUNCTION PREDICTIONS, BETTER PLANT SUPPORT, SYNTENY VIEW AND OTHER IMPROVEMENTS.NUCLEIC ACIDS RESEARCH. VOL. 43. ISSUE D1. P. D240 -D249 | 21 | 50% | 23 |
| 10 | POWELL, S , FORSLUND, K , SZKLARCZYK, D , TRACHANA, K , ROTH, A , HUERTA-CEPAS, J , GABALDON, T , RATTEI, T , CREEVEY, C , KUHN, M , ET AL (2014) EGGNOG V4.0: NESTED ORTHOLOGY INFERENCE ACROSS 3686 ORGANISMS.NUCLEIC ACIDS RESEARCH. VOL. 42. ISSUE D1. P. D231 -D239 | 22 | 35% | 112 |
Classes with closest relation at Level 1 |