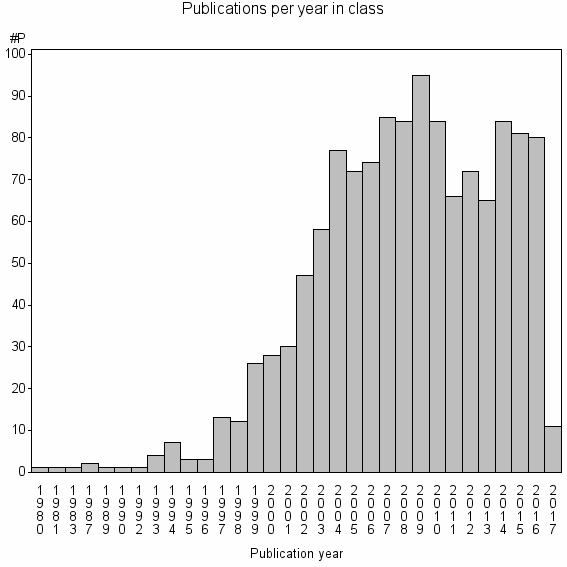
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 8300 | 1269 | 52.9 | 87% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 8300 | 1 | WHOLE GENOME DUPLICATION//GENOME DUPLICATION//GENE DUPLICATION | 1269 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | WHOLE GENOME DUPLICATION | authKW | 617391 | 6% | 36% | 72 |
| 2 | GENOME DUPLICATION | authKW | 350246 | 5% | 23% | 62 |
| 3 | GENE DUPLICATION | authKW | 350126 | 13% | 9% | 169 |
| 4 | PLANT GENOME M PING | address | 163753 | 3% | 17% | 40 |
| 5 | DOSAGE BALANCE | authKW | 153987 | 1% | 80% | 8 |
| 6 | GENE DUPLICABILITY | authKW | 144365 | 0% | 100% | 6 |
| 7 | NEOFUNCTIONALIZATION | authKW | 136828 | 2% | 25% | 23 |
| 8 | PALEOPOLYPLOIDY | authKW | 132324 | 1% | 50% | 11 |
| 9 | SUBFUNCTIONALIZATION | authKW | 128491 | 2% | 21% | 25 |
| 10 | GENE RETENTION | authKW | 128319 | 1% | 67% | 8 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 30451 | 64% | 0% | 807 |
| 2 | Evolutionary Biology | 21710 | 27% | 0% | 348 |
| 3 | Biotechnology & Applied Microbiology | 2363 | 19% | 0% | 244 |
| 4 | Biochemistry & Molecular Biology | 2208 | 36% | 0% | 463 |
| 5 | Mathematical & Computational Biology | 1759 | 7% | 0% | 90 |
| 6 | Developmental Biology | 790 | 5% | 0% | 68 |
| 7 | Plant Sciences | 731 | 12% | 0% | 151 |
| 8 | Biochemical Research Methods | 304 | 6% | 0% | 77 |
| 9 | Biology | 292 | 5% | 0% | 63 |
| 10 | Cell Biology | 106 | 7% | 0% | 86 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PLANT GENOME M PING | 163753 | 3% | 17% | 40 |
| 2 | BIOINFORMAT BIOL STAT | 106991 | 1% | 34% | 13 |
| 3 | PROGRAM COMPARAT EVOLUTIONARY GENOM | 66624 | 0% | 46% | 6 |
| 4 | HUMAN GENOME NETWORK PROJECT | 48122 | 0% | 100% | 2 |
| 5 | INFORMAT GEN STRUCT | 48122 | 0% | 100% | 2 |
| 6 | IPLANT ABORAT | 32738 | 1% | 19% | 7 |
| 7 | MOL MARINE BIOL | 31652 | 0% | 26% | 5 |
| 8 | LEHRSTUHL ZOOL EVOLUTIONSBIOL | 29859 | 0% | 21% | 6 |
| 9 | CAFS AQUAT GENOM | 29608 | 0% | 31% | 4 |
| 10 | SMURFIT GENET | 27747 | 2% | 5% | 23 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR BIOLOGY AND EVOLUTION | 51248 | 9% | 2% | 109 |
| 2 | GENOME BIOLOGY AND EVOLUTION | 24364 | 3% | 3% | 37 |
| 3 | JOURNAL OF EXPERIMENTAL ZOOLOGY PART B-MOLECULAR AND DEVELOPMENTAL EVOLUTION | 21527 | 2% | 4% | 25 |
| 4 | BIOTECHFORUM ( BTF ) | 19628 | 4% | 2% | 46 |
| 5 | BMC EVOLUTIONARY BIOLOGY | 17283 | 4% | 1% | 49 |
| 6 | TRENDS IN GENETICS | 16320 | 3% | 2% | 43 |
| 7 | JOURNAL OF MOLECULAR EVOLUTION | 9554 | 3% | 1% | 39 |
| 8 | GENOME BIOLOGY | 6281 | 2% | 1% | 28 |
| 9 | GENOME RESEARCH | 4451 | 1% | 1% | 16 |
| 10 | BMC GENOMICS | 4376 | 3% | 0% | 42 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | WHOLE GENOME DUPLICATION | 617391 | 6% | 36% | 72 | Search WHOLE+GENOME+DUPLICATION | Search WHOLE+GENOME+DUPLICATION |
| 2 | GENOME DUPLICATION | 350246 | 5% | 23% | 62 | Search GENOME+DUPLICATION | Search GENOME+DUPLICATION |
| 3 | GENE DUPLICATION | 350126 | 13% | 9% | 169 | Search GENE+DUPLICATION | Search GENE+DUPLICATION |
| 4 | DOSAGE BALANCE | 153987 | 1% | 80% | 8 | Search DOSAGE+BALANCE | Search DOSAGE+BALANCE |
| 5 | GENE DUPLICABILITY | 144365 | 0% | 100% | 6 | Search GENE+DUPLICABILITY | Search GENE+DUPLICABILITY |
| 6 | NEOFUNCTIONALIZATION | 136828 | 2% | 25% | 23 | Search NEOFUNCTIONALIZATION | Search NEOFUNCTIONALIZATION |
| 7 | PALEOPOLYPLOIDY | 132324 | 1% | 50% | 11 | Search PALEOPOLYPLOIDY | Search PALEOPOLYPLOIDY |
| 8 | SUBFUNCTIONALIZATION | 128491 | 2% | 21% | 25 | Search SUBFUNCTIONALIZATION | Search SUBFUNCTIONALIZATION |
| 9 | GENE RETENTION | 128319 | 1% | 67% | 8 | Search GENE+RETENTION | Search GENE+RETENTION |
| 10 | HOX CLUSTER | 120904 | 1% | 36% | 14 | Search HOX+CLUSTER | Search HOX+CLUSTER |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | WANG, YP , WANG, XY , PATERSON, AH , (2012) GENOME AND GENE DUPLICATIONS AND GENE EXPRESSION DIVERGENCE: A VIEW FROM PLANTS.YEAR IN EVOLUTIONARY BIOLOGY. VOL. 1256. ISSUE . P. 1-14 | 68 | 61% | 49 |
| 2 | INNAN, H , KONDRASHOV, F , (2010) THE EVOLUTION OF GENE DUPLICATIONS: CLASSIFYING AND DISTINGUISHING BETWEEN MODELS.NATURE REVIEWS GENETICS. VOL. 11. ISSUE 2. P. 97 -108 | 44 | 59% | 408 |
| 3 | SATO, Y , NISHIDA, M , (2010) TELEOST FISH WITH SPECIFIC GENOME DUPLICATION AS UNIQUE MODELS OF VERTEBRATE EVOLUTION.ENVIRONMENTAL BIOLOGY OF FISHES. VOL. 88. ISSUE 2. P. 169 -188 | 75 | 59% | 30 |
| 4 | GLASAUER, SMK , NEUHAUSS, SCF , (2014) WHOLE-GENOME DUPLICATION IN TELEOST FISHES AND ITS EVOLUTIONARY CONSEQUENCES.MOLECULAR GENETICS AND GENOMICS. VOL. 289. ISSUE 6. P. 1045 -1060 | 62 | 48% | 52 |
| 5 | CONANT, GC , BIRCHLER, JA , PIRES, JC , (2014) DOSAGE, DUPLICATION, AND DIPLOIDIZATION: CLARIFYING THE INTERPLAY OF MULTIPLE MODELS FOR DUPLICATE GENE EVOLUTION OVER TIME.CURRENT OPINION IN PLANT BIOLOGY. VOL. 19. ISSUE . P. 91 -98 | 48 | 65% | 40 |
| 6 | BYRNE, KP , BLANC, G , (2006) COMPUTATIONAL ANALYSES OF ANCIENT POLYPLOIDY.CURRENT BIOINFORMATICS. VOL. 1. ISSUE 2. P. 131 -146 | 73 | 66% | 3 |
| 7 | TAYLOR, JS , RAES, J , (2004) DUPLICATION AND DIVERGENCE: THE EVOLUTION OF NEW GENES AND OLD IDEAS.ANNUAL REVIEW OF GENETICS. VOL. 38. ISSUE . P. 615 -643 | 55 | 57% | 397 |
| 8 | JIANG, WK , LIU, YL , XIA, EH , GAO, LZ , (2013) PREVALENT ROLE OF GENE FEATURES IN DETERMINING EVOLUTIONARY FATES OF WHOLE-GENOME DUPLICATION DUPLICATED GENES IN FLOWERING PLANTS.PLANT PHYSIOLOGY. VOL. 161. ISSUE 4. P. 1844-1861 | 53 | 61% | 24 |
| 9 | FREELING, M , (2009) BIAS IN PLANT GENE CONTENT FOLLOWING DIFFERENT SORTS OF DUPLICATION: TANDEM, WHOLE-GENOME, SEGMENTAL, OR BY TRANSPOSITION.ANNUAL REVIEW OF PLANT BIOLOGY. VOL. 60. ISSUE . P. 433-453 | 46 | 55% | 261 |
| 10 | ACHARYA, D , GHOSH, TC , (2016) GLOBAL ANALYSIS OF HUMAN DUPLICATED GENES REVEALS THE RELATIVE IMPORTANCE OF WHOLE-GENOME DUPLICATES ORIGINATED IN THE EARLY VERTEBRATE EVOLUTION.BMC GENOMICS. VOL. 17. ISSUE . P. - | 47 | 71% | 0 |
Classes with closest relation at Level 1 |