Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
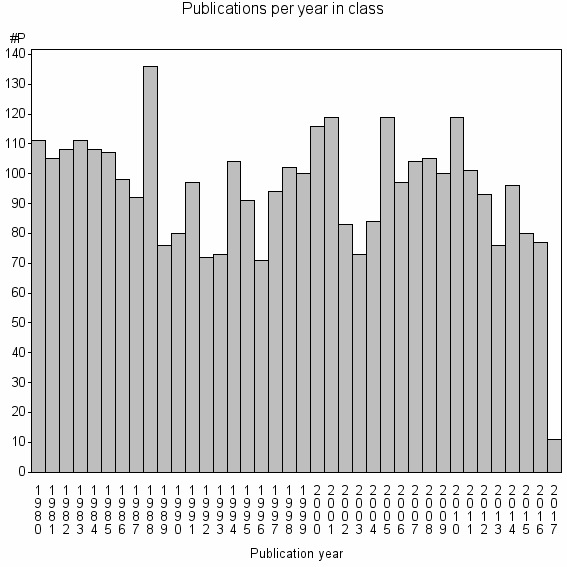
| 360 | 3589 | 42.4 | 66% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 1297 | 2 | RIBOSOME//TRANSLATION TERMINATION//TMRNA | 8599 |
| 360 | 1 | RIBOSOME//PEPTIDYL TRANSFERASE//AG RIBOSOMEN | 3589 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RIBOSOME | authKW | 903018 | 13% | 23% | 462 |
| 2 | PEPTIDYL TRANSFERASE | authKW | 238634 | 1% | 64% | 44 |
| 3 | AG RIBOSOMEN | address | 234628 | 1% | 69% | 40 |
| 4 | EF G | authKW | 170308 | 1% | 69% | 29 |
| 5 | RIBOSOME STRUCTURE | authKW | 160259 | 1% | 61% | 31 |
| 6 | 80S RIBOSOME | authKW | 156350 | 1% | 74% | 25 |
| 7 | MOL BIOL RNA | address | 155833 | 2% | 30% | 61 |
| 8 | PEPTIDYL TRANSFERASE CENTER | authKW | 147928 | 1% | 87% | 20 |
| 9 | ELONGATION FACTOR G | authKW | 131913 | 1% | 57% | 27 |
| 10 | RIBOSOME RECYCLING FACTOR | authKW | 117314 | 1% | 69% | 20 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 41077 | 85% | 0% | 3049 |
| 2 | Biophysics | 5178 | 15% | 0% | 522 |
| 3 | Cell Biology | 3653 | 18% | 0% | 639 |
| 4 | Biochemical Research Methods | 305 | 4% | 0% | 145 |
| 5 | Biology | 291 | 3% | 0% | 117 |
| 6 | Microbiology | 175 | 4% | 0% | 159 |
| 7 | Mathematical & Computational Biology | 19 | 1% | 0% | 27 |
| 8 | Crystallography | 8 | 1% | 0% | 39 |
| 9 | Genetics & Heredity | 5 | 2% | 0% | 75 |
| 10 | Microscopy | 1 | 0% | 0% | 6 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | AG RIBOSOMEN | 234628 | 1% | 69% | 40 |
| 2 | MOL BIOL RNA | 155833 | 2% | 30% | 61 |
| 3 | HLTH INC | 72571 | 0% | 53% | 16 |
| 4 | PHYS BIOCHEM | 55595 | 2% | 9% | 70 |
| 5 | ULTRASTRUKTURNETZWERK | 47098 | 0% | 46% | 12 |
| 6 | RIBOSOMAL ORIGINS EVOLUT | 43744 | 0% | 86% | 6 |
| 7 | PROGRAM CELL MOL DEV BIOL BIOPHYS | 41676 | 0% | 70% | 7 |
| 8 | RNA REGULAT | 40521 | 0% | 53% | 9 |
| 9 | SINSHEIMER S | 36769 | 1% | 13% | 34 |
| 10 | GENOM RNOM | 32653 | 0% | 30% | 13 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF MOLECULAR BIOLOGY | 38827 | 9% | 1% | 327 |
| 2 | RNA | 32089 | 3% | 4% | 98 |
| 3 | MOLECULAR BIOLOGY | 18727 | 3% | 2% | 102 |
| 4 | BIOCHIMIE | 17678 | 3% | 2% | 112 |
| 5 | RNA-A PUBLICATION OF THE RNA SOCIETY | 15975 | 1% | 4% | 49 |
| 6 | NUCLEIC ACIDS RESEARCH | 15410 | 8% | 1% | 270 |
| 7 | NATURE STRUCTURAL & MOLECULAR BIOLOGY | 7204 | 1% | 2% | 43 |
| 8 | BIOCHEMISTRY | 5624 | 5% | 0% | 192 |
| 9 | MOLECULAR CELL | 5516 | 2% | 1% | 60 |
| 10 | FEBS LETTERS | 5342 | 5% | 0% | 163 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RIBOSOME | 903018 | 13% | 23% | 462 | Search RIBOSOME | Search RIBOSOME |
| 2 | PEPTIDYL TRANSFERASE | 238634 | 1% | 64% | 44 | Search PEPTIDYL+TRANSFERASE | Search PEPTIDYL+TRANSFERASE |
| 3 | EF G | 170308 | 1% | 69% | 29 | Search EF+G | Search EF+G |
| 4 | RIBOSOME STRUCTURE | 160259 | 1% | 61% | 31 | Search RIBOSOME+STRUCTURE | Search RIBOSOME+STRUCTURE |
| 5 | 80S RIBOSOME | 156350 | 1% | 74% | 25 | Search 80S+RIBOSOME | Search 80S+RIBOSOME |
| 6 | PEPTIDYL TRANSFERASE CENTER | 147928 | 1% | 87% | 20 | Search PEPTIDYL+TRANSFERASE+CENTER | Search PEPTIDYL+TRANSFERASE+CENTER |
| 7 | ELONGATION FACTOR G | 131913 | 1% | 57% | 27 | Search ELONGATION+FACTOR+G | Search ELONGATION+FACTOR+G |
| 8 | RIBOSOME RECYCLING FACTOR | 117314 | 1% | 69% | 20 | Search RIBOSOME+RECYCLING+FACTOR | Search RIBOSOME+RECYCLING+FACTOR |
| 9 | PEPTIDE BOND FORMATION | 116973 | 1% | 51% | 27 | Search PEPTIDE+BOND+FORMATION | Search PEPTIDE+BOND+FORMATION |
| 10 | MRNA ANALOG | 114604 | 0% | 84% | 16 | Search MRNA+ANALOG | Search MRNA+ANALOG |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | VOORHEES, RM , RAMAKRISHNAN, V , (2013) STRUCTURAL BASIS OF THE TRANSLATIONAL ELONGATION CYCLE.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 82. VOL. 82. ISSUE . P. 203-236 | 146 | 84% | 62 |
| 2 | FRANK, J , (2017) THE TRANSLATION ELONGATION CYCLE-CAPTURING MULTIPLE STATES BY CRYO-ELECTRON MICROSCOPY.PHILOSOPHICAL TRANSACTIONS OF THE ROYAL SOCIETY B-BIOLOGICAL SCIENCES. VOL. 372. ISSUE 1716. P. - | 76 | 82% | 1 |
| 3 | GREEN, R , NOLLER, HF , (1997) RIBOSOMES AND TRANSLATION.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 66. ISSUE . P. 679-716 | 139 | 81% | 321 |
| 4 | AGIRREZABALA, X , VALLE, M , (2015) STRUCTURAL INSIGHTS INTO TRNA DYNAMICS ON THE RIBOSOME.INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. VOL. 16. ISSUE 5. P. 9866 -9895 | 139 | 65% | 5 |
| 5 | SCHMEING, TM , RAMAKRISHNAN, V , (2009) WHAT RECENT RIBOSOME STRUCTURES HAVE REVEALED ABOUT THE MECHANISM OF TRANSLATION.NATURE. VOL. 461. ISSUE 7268. P. 1234 -1242 | 91 | 72% | 302 |
| 6 | LING, C , ERMOLENKO, DN , (2016) STRUCTURAL INSIGHTS INTO RIBOSOME TRANSLOCATION.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 7. ISSUE 5. P. 620 -636 | 92 | 87% | 2 |
| 7 | AGIRREZABALA, X , FRANK, J , (2009) ELONGATION IN TRANSLATION AS A DYNAMIC INTERACTION AMONG THE RIBOSOME, TRNA, AND ELONGATION FACTORS EF-G AND EF-TU.QUARTERLY REVIEWS OF BIOPHYSICS. VOL. 42. ISSUE 3. P. 159-200 | 121 | 77% | 56 |
| 8 | YAMAMOTO, H , QIN, Y , ACHENBACH, J , LI, CM , KIJEK, J , SPAHN, CMT , NIERHAUS, KH , (2014) EF-G AND EF4: TRANSLOCATION AND BACK-TRANSLOCATION ON THE BACTERIAL RIBOSOME.NATURE REVIEWS MICROBIOLOGY. VOL. 12. ISSUE 2. P. 89 -100 | 90 | 87% | 20 |
| 9 | HOLTKAMP, W , WINTERMEYER, W , RODNINA, MV , (2014) SYNCHRONOUS TRNA MOVEMENTS DURING TRANSLOCATION ON THE RIBOSOME ARE ORCHESTRATED BY ELONGATION FACTOR G AND GTP HYDROLYSIS.BIOESSAYS. VOL. 36. ISSUE 10. P. 908 -918 | 83 | 95% | 4 |
| 10 | HOLTKAMP, W , CUNHA, CE , PESKE, F , KONEVEGA, AL , WINTERMEYER, W , RODNINA, MV , (2014) GTP HYDROLYSIS BY EF-G SYNCHRONIZES TRNA MOVEMENT ON SMALL AND LARGE RIBOSOMAL SUBUNITS.EMBO JOURNAL. VOL. 33. ISSUE 9. P. 1073-1085 | 75 | 96% | 15 |
Classes with closest relation at Level 1 |