Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
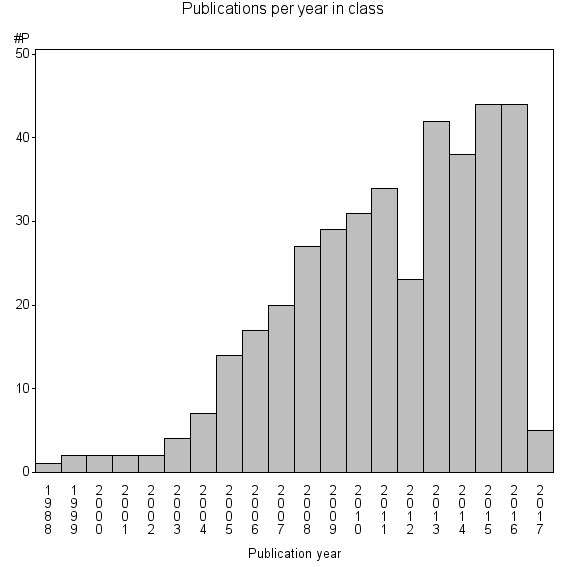
| 22276 | 388 | 45.0 | 88% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 548 | 2 | BMC SYSTEMS BIOLOGY//MATHEMATICAL & COMPUTATIONAL BIOLOGY//BOOLEAN NETWORKS | 14655 |
| 22276 | 1 | SIGNALING SYST//INTRACELLULAR DYNAMIC//NF KAPPA B SIGNAL TRANSDUCTION NETWORK | 388 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | SIGNALING SYST | address | 233689 | 4% | 21% | 14 |
| 2 | INTRACELLULAR DYNAMIC | authKW | 157397 | 1% | 100% | 2 |
| 3 | NF KAPPA B SIGNAL TRANSDUCTION NETWORK | authKW | 157397 | 1% | 100% | 2 |
| 4 | P53 MDM2 NEGATIVE FEEDBACK LOOP | authKW | 157397 | 1% | 100% | 2 |
| 5 | P53 MDM2 SYSTEM | authKW | 157397 | 1% | 100% | 2 |
| 6 | P53 PULSE | authKW | 157397 | 1% | 100% | 2 |
| 7 | PARABOLIC HYPERBOLIC COUPLED MODEL | authKW | 157397 | 1% | 100% | 2 |
| 8 | REPRESSILATORS | authKW | 157397 | 1% | 100% | 2 |
| 9 | QUANTITAT COMPUTAT BIOSCI | address | 122960 | 1% | 31% | 5 |
| 10 | CELL IMAGING | address | 120519 | 3% | 14% | 11 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 7008 | 25% | 0% | 96 |
| 2 | Cell Biology | 1449 | 32% | 0% | 124 |
| 3 | Biology | 910 | 14% | 0% | 55 |
| 4 | Biochemistry & Molecular Biology | 691 | 37% | 0% | 143 |
| 5 | Biochemical Research Methods | 191 | 8% | 0% | 32 |
| 6 | Biophysics | 144 | 8% | 0% | 31 |
| 7 | Genetics & Heredity | 45 | 6% | 0% | 23 |
| 8 | Physics, Mathematical | 42 | 4% | 0% | 14 |
| 9 | Mathematics, Applied | 26 | 5% | 0% | 18 |
| 10 | Biotechnology & Applied Microbiology | 25 | 5% | 0% | 19 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | SIGNALING SYST | 233689 | 4% | 21% | 14 |
| 2 | QUANTITAT COMPUTAT BIOSCI | 122960 | 1% | 31% | 5 |
| 3 | CELL IMAGING | 120519 | 3% | 14% | 11 |
| 4 | BIOPROC DESIGN | 104930 | 1% | 67% | 2 |
| 5 | MATH ONCOL | 83939 | 1% | 27% | 4 |
| 6 | BIOL BIOMED BIOSCI | 78699 | 0% | 100% | 1 |
| 7 | CELL INTERACT GRP | 78699 | 0% | 100% | 1 |
| 8 | COMPUTAT SYST BIOL TRENTO | 78699 | 0% | 100% | 1 |
| 9 | CRBM MACROMOL BIOCHEM | 78699 | 0% | 100% | 1 |
| 10 | DATA ANAL MODELING DYNAM PROC LIFE SCI | 78699 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CELL SYSTEMS | 12337 | 1% | 4% | 4 |
| 2 | IET SYSTEMS BIOLOGY | 11601 | 2% | 2% | 7 |
| 3 | BMC SYSTEMS BIOLOGY | 7050 | 3% | 1% | 12 |
| 4 | MOLECULAR SYSTEMS BIOLOGY | 6646 | 2% | 1% | 8 |
| 5 | IEE PROCEEDINGS SYSTEMS BIOLOGY | 3789 | 1% | 2% | 2 |
| 6 | BIOSYSTEMS | 2895 | 3% | 0% | 10 |
| 7 | JOURNAL OF THEORETICAL BIOLOGY | 2893 | 5% | 0% | 20 |
| 8 | SCIENCE SIGNALING | 2487 | 2% | 0% | 7 |
| 9 | PLOS COMPUTATIONAL BIOLOGY | 2434 | 3% | 0% | 12 |
| 10 | WILEY INTERDISCIPLINARY REVIEWS-SYSTEMS BIOLOGY AND MEDICINE | 2294 | 1% | 1% | 3 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | INTRACELLULAR DYNAMIC | 157397 | 1% | 100% | 2 | Search INTRACELLULAR+DYNAMIC | Search INTRACELLULAR+DYNAMIC |
| 2 | NF KAPPA B SIGNAL TRANSDUCTION NETWORK | 157397 | 1% | 100% | 2 | Search NF+KAPPA+B+SIGNAL+TRANSDUCTION+NETWORK | Search NF+KAPPA+B+SIGNAL+TRANSDUCTION+NETWORK |
| 3 | P53 MDM2 NEGATIVE FEEDBACK LOOP | 157397 | 1% | 100% | 2 | Search P53+MDM2+NEGATIVE+FEEDBACK+LOOP | Search P53+MDM2+NEGATIVE+FEEDBACK+LOOP |
| 4 | P53 MDM2 SYSTEM | 157397 | 1% | 100% | 2 | Search P53+MDM2+SYSTEM | Search P53+MDM2+SYSTEM |
| 5 | P53 PULSE | 157397 | 1% | 100% | 2 | Search P53+PULSE | Search P53+PULSE |
| 6 | PARABOLIC HYPERBOLIC COUPLED MODEL | 157397 | 1% | 100% | 2 | Search PARABOLIC+HYPERBOLIC+COUPLED+MODEL | Search PARABOLIC+HYPERBOLIC+COUPLED+MODEL |
| 7 | REPRESSILATORS | 157397 | 1% | 100% | 2 | Search REPRESSILATORS | Search REPRESSILATORS |
| 8 | GENETIC OSCILLATIONS | 118045 | 1% | 50% | 3 | Search GENETIC+OSCILLATIONS | Search GENETIC+OSCILLATIONS |
| 9 | P53 NETWORK | 88532 | 1% | 38% | 3 | Search P53+NETWORK | Search P53+NETWORK |
| 10 | ACTIVATOR REPRESSOR SYSTEMS | 78699 | 0% | 100% | 1 | Search ACTIVATOR+REPRESSOR+SYSTEMS | Search ACTIVATOR+REPRESSOR+SYSTEMS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SUN, TZ , CUI, J , (2015) DYNAMICS OF P53 IN RESPONSE TO DNA DAMAGE: MATHEMATICAL MODELING AND PERSPECTIVE.PROGRESS IN BIOPHYSICS & MOLECULAR BIOLOGY. VOL. 119. ISSUE 2. P. 175 -182 | 46 | 45% | 1 |
| 2 | CAO, Z , WANG, WD , (2015) ADVANCES IN THE RESEARCH OF P53 PULSES.PROGRESS IN BIOCHEMISTRY AND BIOPHYSICS. VOL. 42. ISSUE 2. P. 147 -153 | 22 | 69% | 1 |
| 3 | ZAMBRANO, S , DE TOMA, I , PIFFER, A , BIANCHI, ME , AGRESTI, A , (2016) NF-KAPPA B OSCILLATIONS TRANSLATE INTO FUNCTIONALLY RELATED PATTERNS OF GENE EXPRESSION.ELIFE. VOL. 5. ISSUE . P. - | 28 | 43% | 3 |
| 4 | LIU, B , YAN, SW , (2011) CELLULAR RESPONSE TO IRRADIATION.COMMUNICATIONS IN THEORETICAL PHYSICS. VOL. 55. ISSUE 5. P. 921-924 | 22 | 71% | 1 |
| 5 | ICHIKAWA, K , OHSHIMA, D , SAGARA, H , (2015) REGULATION OF SIGNAL TRANSDUCTION BY SPATIAL PARAMETERS: A CASE IN NF-KAPPA B OSCILLATION.IET SYSTEMS BIOLOGY. VOL. 9. ISSUE 2. P. 41 -51 | 43 | 31% | 1 |
| 6 | MOTHES, J , BUSSE, D , KOFAHL, B , WOLF, J , (2015) SOURCES OF DYNAMIC VARIABILITY IN NF-KAPPA B SIGNAL TRANSDUCTION: A MECHANISTIC MODEL.BIOESSAYS. VOL. 37. ISSUE 4. P. 452 -462 | 24 | 50% | 6 |
| 7 | BASAK, S , BEHAR, M , HOFFMANN, A , (2012) LESSONS FROM MATHEMATICALLY MODELING THE NF-KAPPA B PATHWAY.IMMUNOLOGICAL REVIEWS. VOL. 246. ISSUE . P. 221-238 | 28 | 42% | 31 |
| 8 | SUN, TZ , CUI, J , (2014) A PLAUSIBLE MODEL FOR BIMODAL P53 SWITCH IN DNA DAMAGE RESPONSE.FEBS LETTERS. VOL. 588. ISSUE 5. P. 815-821 | 24 | 51% | 3 |
| 9 | ZAMBRANO, S , BIANCHI, ME , AGRESTI, A , (2014) HIGH-THROUGHPUT ANALYSIS OF NF-KAPPA B DYNAMICS IN SINGLE CELLS REVEALS BASAL NUCLEAR LOCALIZATION OF NF-KAPPA B AND SPONTANEOUS ACTIVATION OF OSCILLATIONS.PLOS ONE. VOL. 9. ISSUE 3. P. - | 17 | 68% | 8 |
| 10 | GAUDET, S , MILLER-JENSEN, K , (2016) REDEFINING SIGNALING PATHWAYS WITH AN EXPANDING SINGLE-CELL TOOLBOX.TRENDS IN BIOTECHNOLOGY. VOL. 34. ISSUE 6. P. 458 -469 | 27 | 39% | 0 |
Classes with closest relation at Level 1 |