Class information for: |
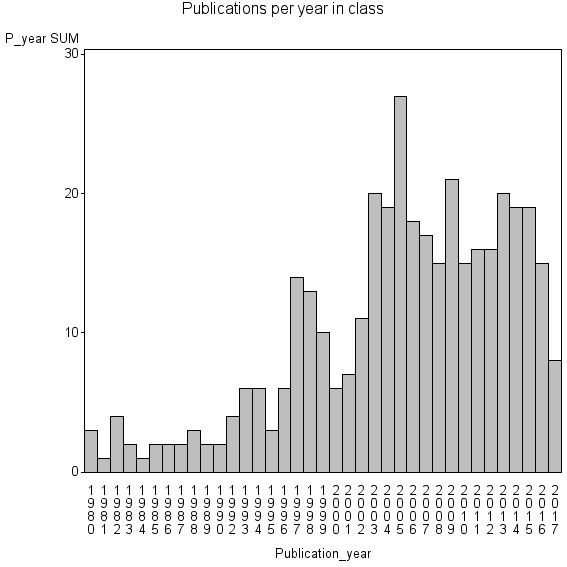
Basic class information |

Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 6 | 4 | CHEMISTRY, ORGANIC//CHEMISTRY, INORGANIC & NUCLEAR//CHEMISTRY, MULTIDISCIPLINARY | 1698077 |
| 156 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 63028 |
| 2461 | 2 | HP MODEL//ENHANCED SAMPLING//JOURNAL OF CHEMICAL THEORY AND COMPUTATION | 4231 |
| 23243 | 1 | HP MODEL//AB OFF LATTICE MODEL//PROTEIN FOLDING PROBLEM | 375 |
Terms with highest relevance score |
| rank | Category | termType | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass |
|---|---|---|---|---|---|---|
| 1 | HP MODEL | authKW | 2206515 | 10% | 69% | 38 |
| 2 | AB OFF LATTICE MODEL | authKW | 420220 | 1% | 100% | 5 |
| 3 | PROTEIN FOLDING PROBLEM | authKW | 336168 | 2% | 50% | 8 |
| 4 | VERTEX SEPARATOR | authKW | 275048 | 2% | 55% | 6 |
| 5 | HYDROPHOBIC POLAR MODEL | authKW | 262634 | 1% | 63% | 5 |
| 6 | FUNCTIONAL MODEL PROTEINS | authKW | 252132 | 1% | 100% | 3 |
| 7 | PULL MOVES | authKW | 252132 | 1% | 100% | 3 |
| 8 | WIDTH BOUNDED SEPARATOR | authKW | 252132 | 1% | 100% | 3 |
| 9 | ANTICOLORING | authKW | 168088 | 1% | 100% | 2 |
| 10 | CSU SOFTWARE | authKW | 168088 | 1% | 100% | 2 |
Web of Science journal categories |
| chi_square_rank | Category | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass |
|---|---|---|---|---|---|
| 1 | Computer Science, Theory & Methods | 1470 | 18% | 0% | 66 |
| 2 | Mathematical & Computational Biology | 1391 | 11% | 0% | 43 |
| 3 | Computer Science, Interdisciplinary Applications | 1378 | 17% | 0% | 63 |
| 4 | Computer Science, Artificial Intelligence | 722 | 12% | 0% | 45 |
| 5 | Physics, Mathematical | 492 | 11% | 0% | 40 |
| 6 | Biochemical Research Methods | 296 | 10% | 0% | 38 |
| 7 | Physics, Fluids & Plasmas | 283 | 7% | 0% | 27 |
| 8 | Mathematics, Applied | 227 | 11% | 0% | 41 |
| 9 | Statistics & Probability | 182 | 6% | 0% | 22 |
| 10 | Biotechnology & Applied Microbiology | 115 | 9% | 0% | 33 |
Address terms |
| chi_square_rank | term | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass |
|---|---|---|---|---|---|
| 1 | PHYS ENGN TANDOGAN | 168088 | 1% | 100% | 2 |
| 2 | GSIT | 126063 | 1% | 50% | 3 |
| 3 | EA 931 | 84044 | 0% | 100% | 1 |
| 4 | GROVER ENGN | 84044 | 0% | 100% | 1 |
| 5 | GRUNDLAGEN INFORMATIONSVERARBEITUNG | 84044 | 0% | 100% | 1 |
| 6 | HUBEI INTELLIGENT INFORMAT PROC REAL TI | 84044 | 0% | 100% | 1 |
| 7 | INT INTEGRATED INTELLIGENT SYST | 84044 | 0% | 100% | 1 |
| 8 | JOHN VO NEUMANN COMP | 84044 | 0% | 100% | 1 |
| 9 | PHYS ENGN BEYTEPE | 84044 | 0% | 100% | 1 |
| 10 | POLYMER PLASMA | 84044 | 0% | 100% | 1 |
Journals |
| chi_square_rank | term | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass |
|---|---|---|---|---|---|
| 1 | JOURNAL OF COMPUTATIONAL BIOLOGY | 18737 | 5% | 1% | 19 |
| 2 | COMPUTATIONAL BIOLOGY AND CHEMISTRY | 4294 | 2% | 1% | 7 |
| 3 | ACTA POLYMERICA SINICA | 2689 | 3% | 0% | 11 |
| 4 | NATURAL COMPUTING | 1914 | 1% | 1% | 3 |
| 5 | LECTURE NOTES IN COMPUTER SCIENCE | 1544 | 11% | 0% | 41 |
| 6 | IEEE TRANSACTIONS ON EVOLUTIONARY COMPUTATION | 1491 | 1% | 0% | 4 |
| 7 | PHYSICAL REVIEW E | 1091 | 7% | 0% | 27 |
| 8 | JOURNAL OF COMBINATORIAL OPTIMIZATION | 993 | 1% | 0% | 4 |
| 9 | INTERDISCIPLINARY SCIENCES-COMPUTATIONAL LIFE SCIENCES | 957 | 1% | 1% | 2 |
| 10 | DISCRETE APPLIED MATHEMATICS | 793 | 2% | 0% | 8 |
Author Key Words |
| chi_square_rank | term | chi_square | shrOfCwithTerm | shrOfTermInClass | termInClass | LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HP MODEL | 2206515 | 10% | 69% | 38 | Search HP+MODEL | Search HP+MODEL |
| 2 | AB OFF LATTICE MODEL | 420220 | 1% | 100% | 5 | Search AB+OFF+LATTICE+MODEL | Search AB+OFF+LATTICE+MODEL |
| 3 | PROTEIN FOLDING PROBLEM | 336168 | 2% | 50% | 8 | Search PROTEIN+FOLDING+PROBLEM | Search PROTEIN+FOLDING+PROBLEM |
| 4 | VERTEX SEPARATOR | 275048 | 2% | 55% | 6 | Search VERTEX+SEPARATOR | Search VERTEX+SEPARATOR |
| 5 | HYDROPHOBIC POLAR MODEL | 262634 | 1% | 63% | 5 | Search HYDROPHOBIC+POLAR+MODEL | Search HYDROPHOBIC+POLAR+MODEL |
| 6 | FUNCTIONAL MODEL PROTEINS | 252132 | 1% | 100% | 3 | Search FUNCTIONAL+MODEL+PROTEINS | Search FUNCTIONAL+MODEL+PROTEINS |
| 7 | PULL MOVES | 252132 | 1% | 100% | 3 | Search PULL+MOVES | Search PULL+MOVES |
| 8 | WIDTH BOUNDED SEPARATOR | 252132 | 1% | 100% | 3 | Search WIDTH+BOUNDED+SEPARATOR | Search WIDTH+BOUNDED+SEPARATOR |
| 9 | ANTICOLORING | 168088 | 1% | 100% | 2 | Search ANTICOLORING | Search ANTICOLORING |
| 10 | CSU SOFTWARE | 168088 | 1% | 100% | 2 | Search CSU+SOFTWARE | Search CSU+SOFTWARE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
Classes with closest relation at Level 1 |