Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
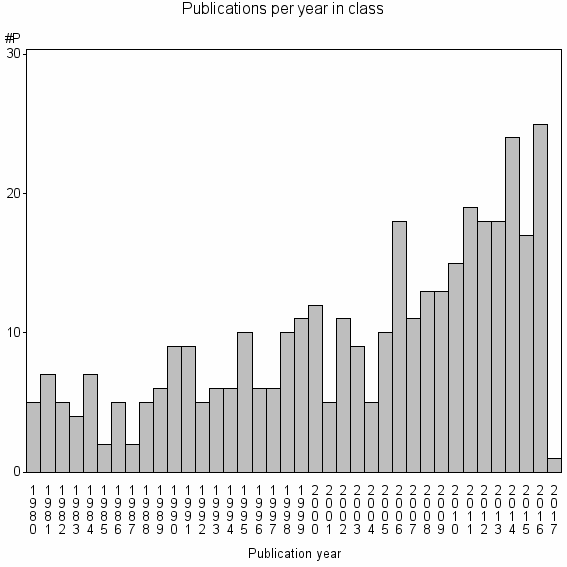
| 22758 | 370 | 41.4 | 77% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 204 | 3 | PROTEASOME//UBIQUITIN//UNFOLDED PROTEIN RESPONSE | 52492 |
| 677 | 2 | PROTEASOME//UBIQUITIN//PROTEASOME INHIBITOR | 13186 |
| 22758 | 1 | ARGINYLATION//JOHANSON BLIZZARD SYNDROME//N END RULE | 370 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | ARGINYLATION | authKW | 2087136 | 8% | 90% | 28 |
| 2 | JOHANSON BLIZZARD SYNDROME | authKW | 1536270 | 6% | 85% | 22 |
| 3 | N END RULE | authKW | 1394772 | 10% | 46% | 37 |
| 4 | N END RULE PATHWAY | authKW | 594187 | 3% | 60% | 12 |
| 5 | L F TRANSFERASE | authKW | 577691 | 2% | 100% | 7 |
| 6 | ATE1 | authKW | 505478 | 2% | 88% | 7 |
| 7 | N DEGRON | authKW | 440140 | 2% | 67% | 8 |
| 8 | N RECOGNIN | authKW | 343862 | 1% | 83% | 5 |
| 9 | ARGINYL TRNA PROTEIN TRANSFERASE | authKW | 330109 | 1% | 100% | 4 |
| 10 | UBR1 GENE | authKW | 330109 | 1% | 100% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 1914 | 59% | 0% | 218 |
| 2 | Cell Biology | 799 | 25% | 0% | 92 |
| 3 | Genetics & Heredity | 143 | 9% | 0% | 35 |
| 4 | Neurosciences | 87 | 11% | 0% | 42 |
| 5 | Biochemical Research Methods | 67 | 5% | 0% | 20 |
| 6 | Pediatrics | 61 | 5% | 0% | 20 |
| 7 | Developmental Biology | 29 | 2% | 0% | 8 |
| 8 | Medical Ethics | 25 | 1% | 0% | 2 |
| 9 | Biophysics | 24 | 4% | 0% | 15 |
| 10 | Mycology | 10 | 1% | 0% | 3 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | KINESIOL PHYS EDUC PHYS PHYSIOL | 165055 | 1% | 100% | 2 |
| 2 | EDINBURGH CRUK CANC | 110035 | 1% | 67% | 2 |
| 3 | CHEM BIOL AREA | 106103 | 1% | 43% | 3 |
| 4 | CEA RECH COR PONDANT 2 | 82527 | 0% | 100% | 1 |
| 5 | CHILDRENS HOSP REBRO | 82527 | 0% | 100% | 1 |
| 6 | DRDC CP CHIM PROT | 82527 | 0% | 100% | 1 |
| 7 | DRDC CS CYTOSQUELETTE | 82527 | 0% | 100% | 1 |
| 8 | GRP PHYSIOL BIOCHEM | 82527 | 0% | 100% | 1 |
| 9 | GRUNENTHAL INNOVAT EARLY CLIN DEV | 82527 | 0% | 100% | 1 |
| 10 | IBMP CNRSUNITE PROPRE RECH 2357 | 82527 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR THERAPY-ONCOLYTICS | 1308 | 0% | 2% | 1 |
| 2 | NEUROCHEMICAL RESEARCH | 1120 | 3% | 0% | 10 |
| 3 | PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA | 602 | 7% | 0% | 27 |
| 4 | JOURNAL OF BIOLOGICAL CHEMISTRY | 553 | 9% | 0% | 33 |
| 5 | CHEMISTRY & BIOLOGY | 489 | 1% | 0% | 4 |
| 6 | NATURE CELL BIOLOGY | 484 | 1% | 0% | 4 |
| 7 | GENES TO CELLS | 383 | 1% | 0% | 3 |
| 8 | JOURNAL OF NEUROCHEMISTRY | 381 | 3% | 0% | 10 |
| 9 | MOLECULAR CELL | 370 | 1% | 0% | 5 |
| 10 | FRONTIERS OF MEDICINE | 368 | 0% | 0% | 1 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | ARGINYLATION | 2087136 | 8% | 90% | 28 | Search ARGINYLATION | Search ARGINYLATION |
| 2 | JOHANSON BLIZZARD SYNDROME | 1536270 | 6% | 85% | 22 | Search JOHANSON+BLIZZARD+SYNDROME | Search JOHANSON+BLIZZARD+SYNDROME |
| 3 | N END RULE | 1394772 | 10% | 46% | 37 | Search N+END+RULE | Search N+END+RULE |
| 4 | N END RULE PATHWAY | 594187 | 3% | 60% | 12 | Search N+END+RULE+PATHWAY | Search N+END+RULE+PATHWAY |
| 5 | L F TRANSFERASE | 577691 | 2% | 100% | 7 | Search L+F+TRANSFERASE | Search L+F+TRANSFERASE |
| 6 | ATE1 | 505478 | 2% | 88% | 7 | Search ATE1 | Search ATE1 |
| 7 | N DEGRON | 440140 | 2% | 67% | 8 | Search N+DEGRON | Search N+DEGRON |
| 8 | N RECOGNIN | 343862 | 1% | 83% | 5 | Search N+RECOGNIN | Search N+RECOGNIN |
| 9 | ARGINYL TRNA PROTEIN TRANSFERASE | 330109 | 1% | 100% | 4 | Search ARGINYL+TRNA+PROTEIN+TRANSFERASE | Search ARGINYL+TRNA+PROTEIN+TRANSFERASE |
| 10 | UBR1 GENE | 330109 | 1% | 100% | 4 | Search UBR1+GENE | Search UBR1+GENE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | TASAKI, T , SRIRAM, SM , PARK, KS , KWON, YT , (2012) THE N-END RULE PATHWAY.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 81. VOL. 81. ISSUE . P. 261-289 | 77 | 55% | 89 |
| 2 | WADAS, B , PIATKOV, KI , BROWER, CS , VARSHAVSKY, A , (2016) ANALYZING N-TERMINAL ARGINYLATION THROUGH THE USE OF PEPTIDE ARRAYS AND DEGRADATION ASSAYS.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 291. ISSUE 40. P. 20976 -20992 | 73 | 72% | 0 |
| 3 | SAHA, S , KASHINA, A , (2011) POSTTRANSLATIONAL ARGINYLATION AS A GLOBAL BIOLOGICAL REGULATOR.DEVELOPMENTAL BIOLOGY. VOL. 358. ISSUE 1. P. 1 -8 | 52 | 83% | 22 |
| 4 | KUMAR, A , BIRNBAUM, MD , PATEL, DM , MORGAN, WM , SINGH, J , BARRIENTOS, A , ZHANG, FL , (2016) POSTTRANSLATIONAL ARGINYLATION ENZYME ATE1 AFFECTS DNA MUTAGENESIS BY REGULATING STRESS RESPONSE.CELL DEATH & DISEASE. VOL. 7. ISSUE . P. - | 40 | 85% | 0 |
| 5 | KASHINA, A , (2014) PROTEIN ARGINYLATION, A GLOBAL BIOLOGICAL REGULATOR THAT TARGETS ACTIN CYTOSKELETON AND THE MUSCLE.ANATOMICAL RECORD-ADVANCES IN INTEGRATIVE ANATOMY AND EVOLUTIONARY BIOLOGY. VOL. 297. ISSUE 9. P. 1630 -1636 | 44 | 79% | 3 |
| 6 | SUKALO, M , FIEDLER, A , GUZMAN, C , SPRANGER, S , ADDOR, MC , MCHEIK, JN , BENAVENT, MO , COBBEN, JM , GILLIS, LA , SHEALY, AG , ET AL (2014) MUTATIONS IN THE HUMAN UBR1 GENE AND THE ASSOCIATED PHENOTYPIC SPECTRUM.HUMAN MUTATION. VOL. 35. ISSUE 5. P. 521-531 | 33 | 85% | 4 |
| 7 | VARSHAVSKY, A , (2011) THE N-END RULE PATHWAY AND REGULATION BY PROTEOLYSIS.PROTEIN SCIENCE. VOL. 20. ISSUE 8. P. 1298 -1345 | 80 | 26% | 155 |
| 8 | GALIANO, MR , GOITEA, VE , HALLAK, ME , (2016) POST-TRANSLATIONAL PROTEIN ARGINYLATION IN THE NORMAL NERVOUS SYSTEM AND IN NEURODEGENERATION.JOURNAL OF NEUROCHEMISTRY. VOL. 138. ISSUE 4. P. 506 -517 | 49 | 51% | 1 |
| 9 | SRIRAM, SM , KIM, BY , KWON, YT , (2011) THE N-END RULE PATHWAY: EMERGING FUNCTIONS AND MOLECULAR PRINCIPLES OF SUBSTRATE RECOGNITION.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 12. ISSUE 11. P. 735 -747 | 55 | 46% | 50 |
| 10 | GRACIET, E , WELLMER, F , (2010) THE PLANT N-END RULE PATHWAY: STRUCTURE AND FUNCTIONS.TRENDS IN PLANT SCIENCE. VOL. 15. ISSUE 8. P. 447-453 | 46 | 59% | 38 |
Classes with closest relation at Level 1 |