Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
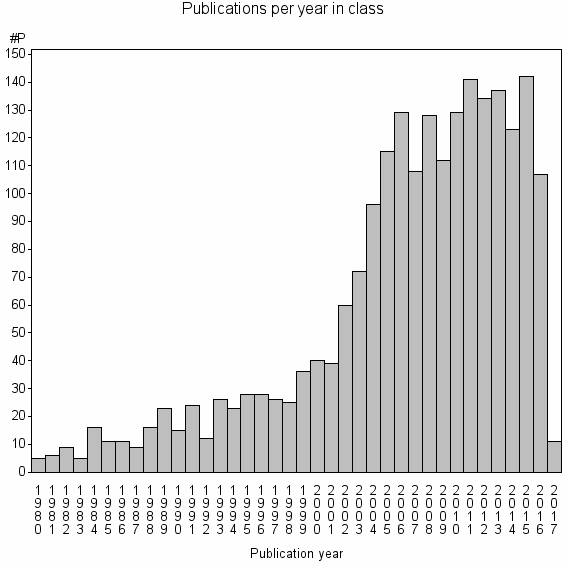
| 2673 | 2177 | 41.3 | 78% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 5 | 4 | CHEMISTRY, ORGANIC//CHEMISTRY, INORGANIC & NUCLEAR//CHEMISTRY, MULTIDISCIPLINARY | 1745167 |
| 183 | 3 | APTAMER//G QUADRUPLEX//RIBOZYME | 56307 |
| 1185 | 2 | RIBOZYME//RNASE P//CATALYTIC RNA | 9289 |
| 2673 | 1 | RNA SECONDARY STRUCTURE//RNA SECONDARY STRUCTURE PREDICTION//PSEUDOKNOT | 2177 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RNA SECONDARY STRUCTURE | authKW | 686510 | 7% | 31% | 156 |
| 2 | RNA SECONDARY STRUCTURE PREDICTION | authKW | 395413 | 2% | 83% | 34 |
| 3 | PSEUDOKNOT | authKW | 348173 | 3% | 35% | 71 |
| 4 | RNA STRUCTURE PREDICTION | authKW | 226251 | 1% | 73% | 22 |
| 5 | NONCODING RNA TECHNOL HLTH | address | 134169 | 2% | 27% | 35 |
| 6 | RNA MOTIF | authKW | 123672 | 1% | 42% | 21 |
| 7 | INTERDISCIPLINARY BIOINFORMAT | address | 95972 | 2% | 14% | 50 |
| 8 | RNA FOLDING | authKW | 95906 | 2% | 13% | 51 |
| 9 | RNA STRUCTURE | authKW | 93559 | 3% | 9% | 72 |
| 10 | NON WATSON CRICK BASE PAIR | authKW | 74792 | 0% | 67% | 8 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 41475 | 25% | 1% | 553 |
| 2 | Biochemical Research Methods | 10755 | 24% | 0% | 519 |
| 3 | Biochemistry & Molecular Biology | 5752 | 44% | 0% | 951 |
| 4 | Biotechnology & Applied Microbiology | 4542 | 20% | 0% | 441 |
| 5 | Computer Science, Interdisciplinary Applications | 2146 | 9% | 0% | 196 |
| 6 | Statistics & Probability | 682 | 5% | 0% | 105 |
| 7 | Biology | 579 | 5% | 0% | 115 |
| 8 | Computer Science, Theory & Methods | 456 | 5% | 0% | 101 |
| 9 | Biophysics | 247 | 5% | 0% | 108 |
| 10 | Mathematics, Interdisciplinary Applications | 95 | 2% | 0% | 43 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NONCODING RNA TECHNOL HLTH | 134169 | 2% | 27% | 35 |
| 2 | INTERDISCIPLINARY BIOINFORMAT | 95972 | 2% | 14% | 50 |
| 3 | GRP BIOINFORMAT COMPUTAT BIOL | 66814 | 0% | 53% | 9 |
| 4 | BIOINFORMAT GRP | 66210 | 4% | 6% | 79 |
| 5 | GRP BCB | 44877 | 0% | 80% | 4 |
| 6 | IBHV | 42928 | 1% | 22% | 14 |
| 7 | AMIB TEAM PROJECT | 42074 | 0% | 100% | 3 |
| 8 | CCNSB | 36056 | 0% | 43% | 6 |
| 9 | IMAGE PROC SECT | 32053 | 0% | 57% | 4 |
| 10 | BIOINFORMAT COMPUTAT BIOL GRP | 29212 | 0% | 42% | 5 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOINFORMATICS | 44454 | 8% | 2% | 184 |
| 2 | JOURNAL OF COMPUTATIONAL BIOLOGY | 40306 | 3% | 4% | 67 |
| 3 | RNA | 37100 | 4% | 3% | 82 |
| 4 | NUCLEIC ACIDS RESEARCH | 35378 | 15% | 1% | 316 |
| 5 | ALGORITHMS FOR MOLECULAR BIOLOGY | 33453 | 1% | 9% | 26 |
| 6 | COMPUTER APPLICATIONS IN THE BIOSCIENCES | 23392 | 2% | 4% | 39 |
| 7 | RNA-A PUBLICATION OF THE RNA SOCIETY | 21277 | 2% | 3% | 44 |
| 8 | BMC BIOINFORMATICS | 19317 | 5% | 1% | 102 |
| 9 | IEEE-ACM TRANSACTIONS ON COMPUTATIONAL BIOLOGY AND BIOINFORMATICS | 12647 | 1% | 3% | 32 |
| 10 | JOURNAL OF BIOINFORMATICS AND COMPUTATIONAL BIOLOGY | 7484 | 1% | 4% | 14 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA SECONDARY STRUCTURE | 686510 | 7% | 31% | 156 | Search RNA+SECONDARY+STRUCTURE | Search RNA+SECONDARY+STRUCTURE |
| 2 | RNA SECONDARY STRUCTURE PREDICTION | 395413 | 2% | 83% | 34 | Search RNA+SECONDARY+STRUCTURE+PREDICTION | Search RNA+SECONDARY+STRUCTURE+PREDICTION |
| 3 | PSEUDOKNOT | 348173 | 3% | 35% | 71 | Search PSEUDOKNOT | Search PSEUDOKNOT |
| 4 | RNA STRUCTURE PREDICTION | 226251 | 1% | 73% | 22 | Search RNA+STRUCTURE+PREDICTION | Search RNA+STRUCTURE+PREDICTION |
| 5 | RNA MOTIF | 123672 | 1% | 42% | 21 | Search RNA+MOTIF | Search RNA+MOTIF |
| 6 | RNA FOLDING | 95906 | 2% | 13% | 51 | Search RNA+FOLDING | Search RNA+FOLDING |
| 7 | RNA STRUCTURE | 93559 | 3% | 9% | 72 | Search RNA+STRUCTURE | Search RNA+STRUCTURE |
| 8 | NON WATSON CRICK BASE PAIR | 74792 | 0% | 67% | 8 | Search NON+WATSON+CRICK+BASE+PAIR | Search NON+WATSON+CRICK+BASE+PAIR |
| 9 | K NONCROSSING RNA STRUCTURE | 70123 | 0% | 100% | 5 | Search K+NONCROSSING+RNA+STRUCTURE | Search K+NONCROSSING+RNA+STRUCTURE |
| 10 | NEAREST NEIGHBOR PARAMETERS | 70123 | 0% | 100% | 5 | Search NEAREST+NEIGHBOR+PARAMETERS | Search NEAREST+NEIGHBOR+PARAMETERS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | LORENZ, R , BERNHART, SH , SIEDERDISSEN, CHZ , TAFER, H , FLAMM, C , STADLER, PF , HOFACKER, IL , (2011) VIENNARNA PACKAGE 2.0.ALGORITHMS FOR MOLECULAR BIOLOGY. VOL. 6. ISSUE . P. - | 64 | 80% | 454 |
| 2 | LORENZ, R , WOLFINGER, MT , TANZER, A , HOFACKER, IL , (2016) PREDICTING RNA SECONDARY STRUCTURES FROM SEQUENCE AND PROBING DATA.METHODS. VOL. 103. ISSUE . P. 86 -98 | 92 | 83% | 3 |
| 3 | GE, P , ZHANG, SJ , (2015) COMPUTATIONAL ANALYSIS OF RNA STRUCTURES WITH CHEMICAL PROBING DATA.METHODS. VOL. 79-80. ISSUE . P. 60 -66 | 78 | 68% | 9 |
| 4 | ANDRONESCU, M , CONDON, A , HOOS, HH , MATHEWS, DH , MURPHY, KP , (2010) COMPUTATIONAL APPROACHES FOR RNA ENERGY PARAMETER ESTIMATION.RNA. VOL. 16. ISSUE 12. P. 2304 -2318 | 79 | 74% | 34 |
| 5 | KWOK, CK , TANG, Y , ASSMANN, SM , BEVILACQUA, PC , (2015) THE RNA STRUCTUROME: TRANSCRIPTOME-WIDE STRUCTURE PROBING WITH NEXT-GENERATION SEQUENCING.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 40. ISSUE 4. P. 221 -232 | 55 | 71% | 24 |
| 6 | PUTON, T , KOZLOWSKI, LP , ROTHER, KM , BUJNICKI, JM , (2013) COMPARNA: A SERVER FOR CONTINUOUS BENCHMARKING OF AUTOMATED METHODS FOR RNA SECONDARY STRUCTURE PREDICTION.NUCLEIC ACIDS RESEARCH. VOL. 41. ISSUE 7. P. 4307-4323 | 58 | 85% | 20 |
| 7 | MATHEWS, DH , SABINA, J , ZUKER, M , TURNER, DH , (1999) EXPANDED SEQUENCE DEPENDENCE OF THERMODYNAMIC PARAMETERS IMPROVES PREDICTION OF RNA SECONDARY STRUCTURE.JOURNAL OF MOLECULAR BIOLOGY. VOL. 288. ISSUE 5. P. 911 -940 | 49 | 53% | 2599 |
| 8 | SHI, YZ , WU, YY , WANG, FH , TAN, ZJ , (2014) RNA STRUCTURE PREDICTION: PROGRESS AND PERSPECTIVE.CHINESE PHYSICS B. VOL. 23. ISSUE 7. P. - | 75 | 68% | 5 |
| 9 | CAPRIOTTI, E , MARTI-RENOM, MA , (2008) COMPUTATIONAL RNA STRUCTURE PREDICTION.CURRENT BIOINFORMATICS. VOL. 3. ISSUE 1. P. 32 -45 | 86 | 68% | 16 |
| 10 | MACHADO-LIMA, A , DEL PORTILLO, HA , DURHAM, AM , (2008) COMPUTATIONAL METHODS IN NONCODING RNA RESEARCH.JOURNAL OF MATHEMATICAL BIOLOGY. VOL. 56. ISSUE 1-2. P. 15-49 | 80 | 71% | 24 |
Classes with closest relation at Level 1 |