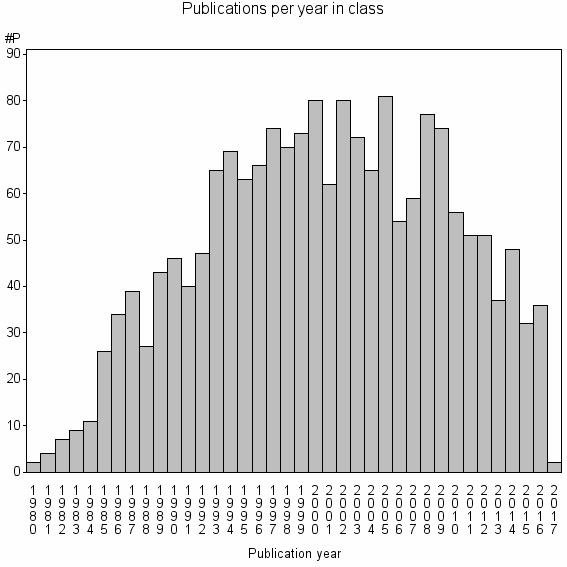
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 4185 | 1832 | 45.2 | 85% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 5 | 4 | CHEMISTRY, ORGANIC//CHEMISTRY, INORGANIC & NUCLEAR//CHEMISTRY, MULTIDISCIPLINARY | 1745167 |
| 183 | 3 | APTAMER//G QUADRUPLEX//RIBOZYME | 56307 |
| 1185 | 2 | RIBOZYME//RNASE P//CATALYTIC RNA | 9289 |
| 4185 | 1 | GROUP I INTRON//RIBOZYME//SELF SPLICING | 1832 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | GROUP I INTRON | authKW | 1885523 | 11% | 57% | 197 |
| 2 | RIBOZYME | authKW | 812858 | 14% | 19% | 256 |
| 3 | SELF SPLICING | authKW | 767983 | 3% | 79% | 58 |
| 4 | GROUP II INTRON | authKW | 736095 | 5% | 46% | 97 |
| 5 | RNA FOLDING | authKW | 456149 | 6% | 27% | 102 |
| 6 | TRANS SPLICING RIBOZYME | authKW | 299988 | 1% | 100% | 18 |
| 7 | GRP ECOL GENET | address | 267148 | 1% | 70% | 23 |
| 8 | RNA REPLACEMENT | authKW | 249990 | 1% | 100% | 15 |
| 9 | GROUP I RIBOZYME | authKW | 220576 | 1% | 88% | 15 |
| 10 | INTRON MOBILITY | authKW | 158006 | 1% | 59% | 16 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 16423 | 76% | 0% | 1389 |
| 2 | Cell Biology | 1442 | 16% | 0% | 292 |
| 3 | Genetics & Heredity | 815 | 10% | 0% | 184 |
| 4 | Biophysics | 656 | 8% | 0% | 144 |
| 5 | Evolutionary Biology | 197 | 3% | 0% | 47 |
| 6 | Biochemical Research Methods | 134 | 4% | 0% | 70 |
| 7 | Microbiology | 127 | 5% | 0% | 91 |
| 8 | Mycology | 41 | 1% | 0% | 14 |
| 9 | Biology | 41 | 2% | 0% | 38 |
| 10 | Biotechnology & Applied Microbiology | 23 | 3% | 0% | 57 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GRP ECOL GENET | 267148 | 1% | 70% | 23 |
| 2 | TC JENKINS BIOPHYS | 81601 | 2% | 15% | 32 |
| 3 | SECT MOL GENET MICROBIOL | 72590 | 3% | 8% | 52 |
| 4 | BECKMAN B400 | 47998 | 1% | 26% | 11 |
| 5 | NANOSENSOR BIOTECHNOL | 41680 | 1% | 10% | 26 |
| 6 | SYNCHROTRON BIOSCI | 33455 | 1% | 13% | 15 |
| 7 | NUCL MAGNET ONANCE MADISON | 33332 | 0% | 100% | 2 |
| 8 | SCI PLANTES PARIS SACLAY IPS2 | 33332 | 0% | 100% | 2 |
| 9 | RNA GRP | 28598 | 1% | 11% | 15 |
| 10 | GENET CELLULAR THER IES | 22208 | 0% | 17% | 8 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA | 43046 | 4% | 3% | 81 |
| 2 | RNA-A PUBLICATION OF THE RNA SOCIETY | 34008 | 3% | 4% | 51 |
| 3 | JOURNAL OF MOLECULAR BIOLOGY | 15337 | 8% | 1% | 147 |
| 4 | NUCLEIC ACIDS RESEARCH | 13678 | 10% | 0% | 181 |
| 5 | NATURE STRUCTURAL BIOLOGY | 8246 | 2% | 2% | 28 |
| 6 | BIOCHEMISTRY | 7650 | 9% | 0% | 158 |
| 7 | MOBILE DNA | 3910 | 0% | 4% | 6 |
| 8 | CURRENT GENETICS | 3020 | 1% | 1% | 26 |
| 9 | CELL | 2937 | 3% | 0% | 52 |
| 10 | CURRENT OPINION IN STRUCTURAL BIOLOGY | 2341 | 1% | 1% | 18 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GROUP I INTRON | 1885523 | 11% | 57% | 197 | Search GROUP+I+INTRON | Search GROUP+I+INTRON |
| 2 | RIBOZYME | 812858 | 14% | 19% | 256 | Search RIBOZYME | Search RIBOZYME |
| 3 | SELF SPLICING | 767983 | 3% | 79% | 58 | Search SELF+SPLICING | Search SELF+SPLICING |
| 4 | GROUP II INTRON | 736095 | 5% | 46% | 97 | Search GROUP+II+INTRON | Search GROUP+II+INTRON |
| 5 | RNA FOLDING | 456149 | 6% | 27% | 102 | Search RNA+FOLDING | Search RNA+FOLDING |
| 6 | TRANS SPLICING RIBOZYME | 299988 | 1% | 100% | 18 | Search TRANS+SPLICING+RIBOZYME | Search TRANS+SPLICING+RIBOZYME |
| 7 | RNA REPLACEMENT | 249990 | 1% | 100% | 15 | Search RNA+REPLACEMENT | Search RNA+REPLACEMENT |
| 8 | GROUP I RIBOZYME | 220576 | 1% | 88% | 15 | Search GROUP+I+RIBOZYME | Search GROUP+I+RIBOZYME |
| 9 | INTRON MOBILITY | 158006 | 1% | 59% | 16 | Search INTRON+MOBILITY | Search INTRON+MOBILITY |
| 10 | TETRALOOP RECEPTOR | 151508 | 1% | 91% | 10 | Search TETRALOOP+RECEPTOR | Search TETRALOOP+RECEPTOR |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | PYLE, AM , (2016) GROUP II INTRON SELF-SPLICING.ANNUAL REVIEW OF BIOPHYSICS, VOL 45. VOL. 45. ISSUE . P. 183 -205 | 109 | 88% | 1 |
| 2 | LAMBOWITZ, AM , ZIMMERLY, S , (2011) GROUP II INTRONS: MOBILE RIBOZYMES THAT INVADE DNA.COLD SPRING HARBOR PERSPECTIVES IN BIOLOGY. VOL. 3. ISSUE 8. P. - | 112 | 76% | 27 |
| 3 | LONNBERG, T , (2011) UNDERSTANDING CATALYSIS OF PHOSPHATE-TRANSFER REACTIONS BY THE LARGE RIBOZYMES.CHEMISTRY-A EUROPEAN JOURNAL. VOL. 17. ISSUE 26. P. 7140 -7153 | 137 | 66% | 3 |
| 4 | ZIMMERLY, S , SEMPER, C , (2015) EVOLUTION OF GROUP II INTRONS.MOBILE DNA. VOL. 6. ISSUE . P. - | 111 | 58% | 17 |
| 5 | PYLE, AM , (2010) THE TERTIARY STRUCTURE OF GROUP II INTRONS: IMPLICATIONS FOR BIOLOGICAL FUNCTION AND EVOLUTION.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 45. ISSUE 3. P. 215-232 | 88 | 83% | 50 |
| 6 | MCNEIL, BA , SEMPER, C , ZIMMERLY, S , (2016) GROUP II INTRONS: VERSATILE RIBOZYMES AND RETROELEMENTS.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 7. ISSUE 3. P. 341 -355 | 73 | 80% | 3 |
| 7 | LEHMANN, K , SCHMIDT, U , (2003) GROUP II INTRONS: STRUCTURE AND CATALYTIC VERSATILITY OF LARGE NATURAL RIBOZYMES.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 38. ISSUE 3. P. 249-303 | 155 | 54% | 103 |
| 8 | LAMBOWITZ, AM , ZIRNMERLY, S , (2004) MOBILE GROUP II INTRONS.ANNUAL REVIEW OF GENETICS. VOL. 38. ISSUE . P. 1-35 | 86 | 79% | 238 |
| 9 | FEDOROVA, O , ZINGLER, N , (2007) GROUP II INTRONS: STRUCTURE, FOLDING AND SPLICING MECHANISM.BIOLOGICAL CHEMISTRY. VOL. 388. ISSUE 7. P. 665 -678 | 98 | 76% | 55 |
| 10 | TORO, N , JIMENEZ-ZURDO, JI , GARCIA-RODRIGUEZ, FM , (2007) BACTERIAL GROUP II INTRONS: NOT JUST SPLICING.FEMS MICROBIOLOGY REVIEWS. VOL. 31. ISSUE 3. P. 342-358 | 83 | 87% | 38 |
Classes with closest relation at Level 1 |