19 Feb. - Summary of preliminary results from wastewater analysis for tracing SARS-CoV-2
Stockholm region, February 19, 2021
Background and method
Sampling of wastewater has been done since mid-April 2020 at Bromma, Henriksdal, and Käppala wastewater treatment plants (WWTP). These treatment plants receive wastewater from a population of approximately 360,000; 860,000 and 500,000, respectively.
The sampling at Henriksdal and Bromma started in week no. 16 (13 April) and the Käppala sampling started in week no. 18, following a sampling protocol developed by KTH. The flow-proportional pooled samples were taken bi-weekly until end of July. From week no.35 the sampling was done every week. After concentration, filtering and preparation, the samples have been analyzed using qPCR technique for genetic material (RNA) belonging to the virus SARS-CoV-2, known to cause the COVID-19 pandemic.
When analyses have not been possible immediately, raw wastewater has been frozen at –20 degrees, and concentrated wastewater or purified RNA stored at -80 C, until the next analysis step has been carried out.
During June and July, KTH researchers have compared four different concentration methods, two of them are commonly used internationally and the other two were modified methods adapted by the KTH team. The sensitivity of two ultrafiltration-based methods and two adsorption and extraction-based methods were compared for the SARS-CoV-2 as well as for two reference viruses. Our investigation concluded that the double ultrafiltration method adapted by KTH has a significantly higher efficiency compared to single filtration and adsorption methods. The scientific article has been published in Science of Total Environment, an internationally leading scientific journal, and is available on this link.
We wish to re-iterate that water and wastewater are not known to be important pathways for transmission of the virus. We have not detected any virus in treated outgoing water and the tracing of SARS-CoV-2 in wastewater should not cause any need for additional precautions for water consumers.
Preliminary Results from SARS-CoV-2 monitoring in Stockholm Region
The RNA signal detected in the wastewater analyses reflects the amount of virus excreted from infected persons within the catchment area, and may thus be used as an indicator when assessing the trend of the COVID-19 pandemic among the population. The analysis results have been converted into gene copy numbers.
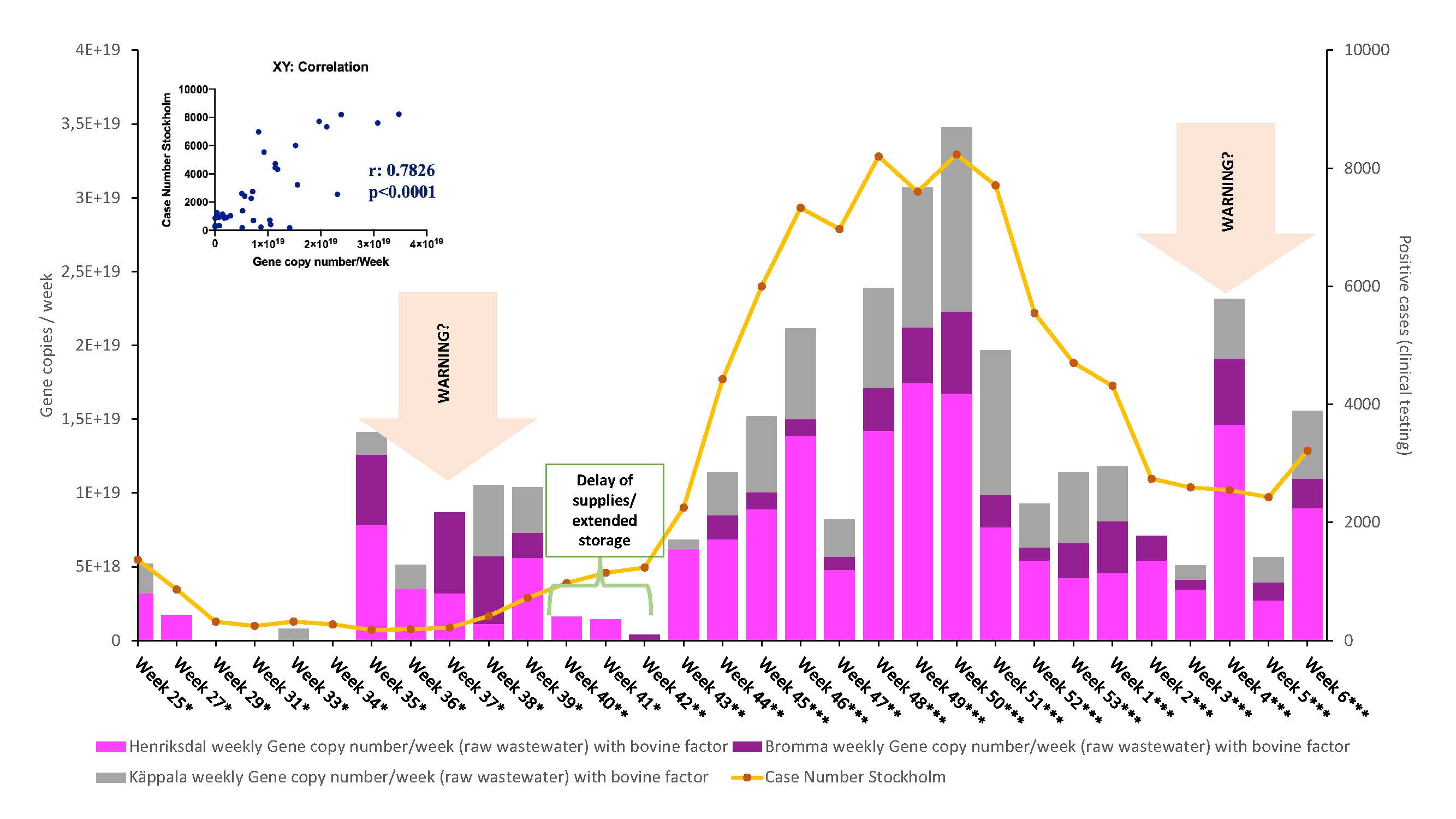
- According to our results, there was an increasing trend between week 35 and 50.
- The viral load in wastewater reached to the highest point in week 50 from the beginning of the pandemic.
- After week 50, the gene copy numbers in wastewater samples started to decrease and significant decreased was detected in week 2.
- Our results correlate relatively well with reported positive case numbers in Stockholm.
- In 2021,
- The virus levels decreased rapidly in week 2, to borderline detection level.
- Especially low levels were found in Bromma and Käppala inlets.
- We detected a significant increase in week 4. Even the gene copy numbers decreased in week 5, an increasing trend was detected in the samples of week 6.
- The results between week 25 (2020) and week 6 (2021) are highly correlated to positive case numbers (p<0.0001).
- We continue to collect triplicate samples from three WWTPs and to perform analysis weekly on fresh samples.
- We also welcome the continued collaboration with Stockholm Vatten och Avfall AB, Käppalaverket, SciLifeLab, Uppsala University and SLU, and international partners, in developing knowledge and methods for wastewater-based epidemiology.
Zeynep Cetecioglu Gurol, Associate Professor
Cecilia Williams, Professor
KTH Royal Institute of Technology
