My Research
Ongoing research
Benchmarking, profiling and optimizing molecular dynamics and molecular docking codes for exascale computing facilities
The computing speed is growing every year and soon we will have access to exascale computing facilities (which can perform 10**18 operations per second). However, if we look back and see whether are there any softwares for doing molecular docking, molecular dynamics or electronic structure theory calculations in connection to many life science projects that can run efficiently on such computing facilities, we do not have any satisfactory answer. It is high time to optimize the codes for running in exascale computing facilities so that we can make full use of the achievements made in hardware developments. My current research is to contribute to this area and I am involved in profiling and benchmarking studies about the performance of popular molecular dynamics software, Gromacs. Once we identify the hotspots, the plan is to optimize the codes so that they can be run on next generation computers.
Research work during 2007-2020 during my association with CBH school
At KTH, initially, my research interests were to investiage the solvent and protein confinement effect on molecular structure and optical properties. I have also studied to some exent the pH dependent and metal-ion specific one photon and two photon properties of organic molecules which have potential applications as diagnostic/imaging agents. After my appointment as a Docent in theoretical chemistry and biology (April 2015) in TheochemBIO, KTH, I am mostly working on computational design of tracers for medical imaging of internal organs such as brain to facilitate early diagnosis of various neurodegenerative diseases. I validate currently available computational methods for their ability to predict various drug-like properties and develop multiscale methods for predicting these properties accurately and reliably. Below, the workflows for multiscale methods for modeling the optical properties of molecules when they are bound to biomolecules like fibrils, membranes, DNA and enzymes and for computing the binding free energies for small molecules with their targets are shown.
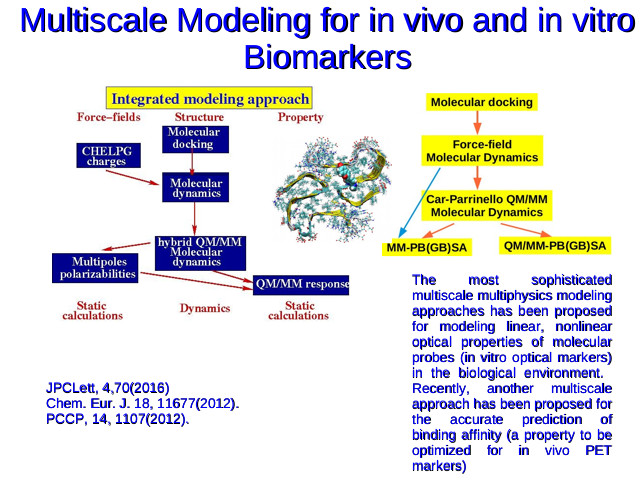
To summarize I can say that my current research activities are around the method development for doing rational medicinal chemistry. I am working on developing hybrid QM/MM and QM fragmentation based multiscale approaches to predict various properties such as binding affinity and binding specificity which are of relevance for therapeutics and diagnostics. I also work on validating the currently available computational methods in predicting the binding affinity for small tracers molecules and drug molecules with biomacromolecules which are potential targets in various neurodegenerative diseases.
In silico design of PET tracers for Alzheimer’s diagnosis:
The objective is to develop improved PET tracers for amyloid and tau imaging to facilitate the diagnosis of Alzheimer’s disease in its early stage. Currently there exist a number of tracers for sensing fibrils. How ever they often lack specificity towards fibrils of amyloid beta peptides, tau protein, amylin and synuclein proteins making them not suitable to distinguish between different types of dementia. There is increasing demand for developing the dementia specific diagnostic molecules which can bind selectively to specific biostructures. In order to improve the specificity towards selective target structures, it is necessary to have understanding at the molecular level and to know details about the mode of binding and the nature of interactions between the biostructures and PET tracers. Computational studies using molecular docking, molecular dynamics and free energy calculations can provide valuable information about various aspects of the interaction between the fibril-PET tracer complexes. Our aim is to study how various currently available PET tracers interact with amyloid beta and tau proteins and based on this knowledge we aim to propose to design principles for the PET tracers which can specifically interact with core sites of amyloid and tau fibrils. Initially, the computational approaches will be tested for their ability to reproduce the available experimental results and once the method is bench-marked it will be used for the design purpose. Within this project, we have also proposed strategies to predict the binding free energies more accurately by incorporating methods based on QM/MM framework. The validation of these approaches in reproducing the experimental binding affinities of various PET tracers to amyloid beta (1-42) fibrils is in progress.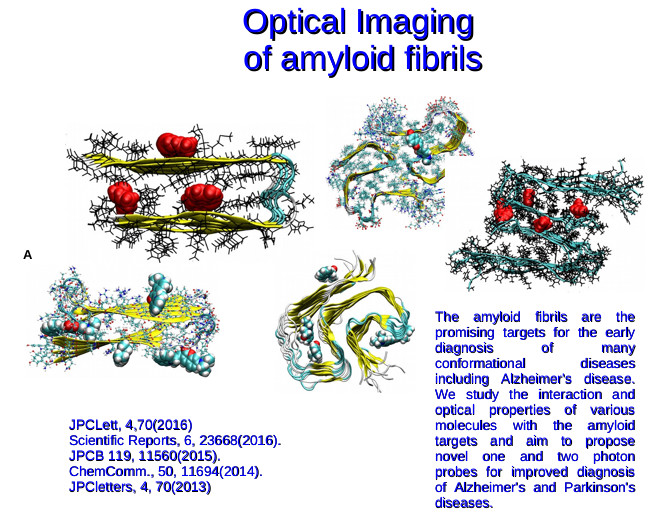
Two photon probes for bio-medical imaging applications:
Design of a new diagnostic probe in health-care research is enormously expensive and time-taking. Moreover, the currently available diagnostic agents are radiation hazardous and so there is need for alternative agents for diagnosis. I also contribute to this subject by studying the working mechanism of various molecular probes and to establish possible structure-property relationships in them for further to be used for designing novel bio-compatible diagnostic probes. In particular, we focus on the two photon probes which can be excited through simultaneous absorption of two photons of low energy (often in the IR region) and so in this two photon microscopy based imaging technique the human body is not exposed to any harmful radiation such as UV, gamma and X-rays. The larger penetrating power of IR radiation helps in imaging the organs located deeper in human-body. For practical applications, the probes should possess large magnitude for two photon absorption (TPA) cross sections which can be computed using quadratic response time dependent density functional theory. In order to model the probes in the realistic biological environment, we have employed an integrated approach that uses molecular docking, molecular dynamics (MD) and hybrid quantum mechanics/molecular mechanics (QM/MM) Car-Parrinello molecular dynamics and hybrid QM/MM response methods that effectively describes different aspects in the probe-target bio-structure system. Using this approach the two photon probes that can be used for in-vivo sensing of metal ions and pH and to follow the membrane disruption have been studied successfully. The mechanism for the decrease (turn-off) or enhancement (turn-on) in TPA due to biological and physiological condition has been established. It is important to recall that the hybrid QM/MM approach that we use as a part of the integrated approach has attracted the Noble prize in chemistry for year-2013 due to their ability to model the reaction mechanism of enzymes and spectroscopy of chromophores within proteins.
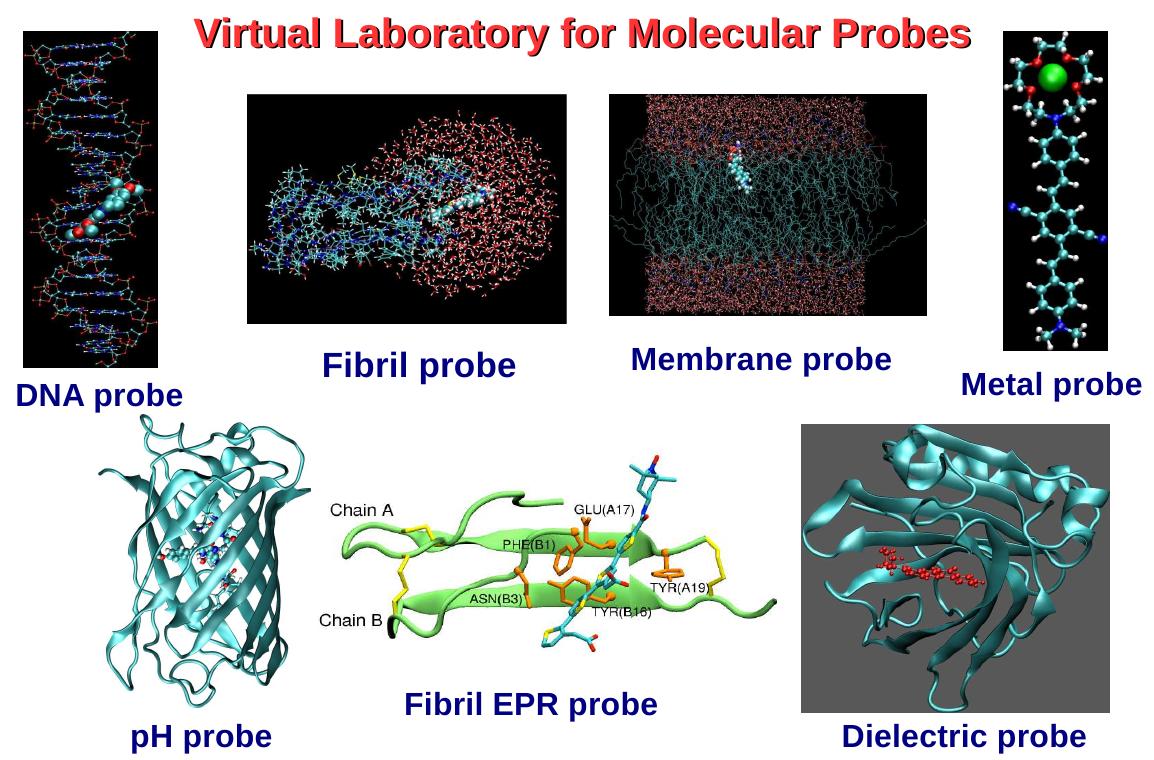
Design of disease-specific biomarkers My research at KTH also covers the design of bio-markers for early-diagnosis of various diseases. This is based on the idea that the diseased state of body is usually associated with (i) change in the conformational nature of proteins (leading to fibril formation) (ii) membranes disruption (iii) abnormal mutations in DNA, (iv) aberrant expression of specific enzymes and (v) deviation from the normal pH values and normal distribution of metal ions and so any molecules that are sensitive to the aforementioned changes in bio-structure and pH and metal ion-distribution, can be used as a diagnostic probe. We investigate various biomarkers that can target bio-structures such as fibril, enzyme active sites, DNA and membranes and can sense pH and specific metal-ions. We study various aspects of (target) biostructure-probe complex system such as the (i) binding modes of probe into the target (ii) probe-induced structural changes in the target (iii) bio-environment induced structural changes in the probe and (iv) bio-environment dependent properties of probes. We have carried out detailed investigation on some of the existing molecular probes to understand their working mechanism. To list a few nile red as a dielectric probe, Thioflavin-T as fibril probe, NAAA as a membrane probe, distyryl derivative as metal probe and methoxy naphthalene derivative as pH probe have been studied. The long-term goal of this project is to propose the structural-motifs for the bio-markers that can be used to diagnose a specific disease.
Tailoring protein backbone through mutations to fine-tune optical properties of GFPs The discovery of green fluorescent proteins and the possibility to integrate it into any parts of hu- man body have revolutionized in vivo sensing of pH and metal ions and imaging of different organs. The light absorption behavior of light in the near UV region and small magnitude for TPA cross section of GFPs limit the practical imaging applications using them and so fine-tuning of one and two photon absorption spectra become necessary. A number of mutation studies exist in literature which contribute to the subject of mutations-induced changes in the one and two photon absorp- tion properties of GFPs. Computationally such relationship between mutation in protein backbone and chromophore optical behavior can be established in a more systematic way and the ”favorable” mutations can be identified. The goal of this project is to derive generalized polarizable force-field for GFPs to be used in the time-dependent density functional theory/molecular mechanics (TD- DFT/MM) methods for computing optical properties and to investigate the mutation-induced fine-tuning of one and two photon optical properties. We aim to come-up with design GFPs with larger TPA cross section for practical medical-imaging applications.
Research work during Ph.D at Indian Institute of Science, Bangalore, India
I have started my research career as a Ph. D student in computational materials science (in Indian Institute of Science, Bangalore, India from 1998) and I was studying the temperature and pressure effect on organic molecular crystals. In particular I addressed the effect of these external variables on cell parameters, molecular packing, intramolecular structure and spectroscopic properties. I have developed Monte Carlo code for doing simulations in isothermal-isobaric ensemble which also accounted for sampling over certain larger amplitude torsional modes and vibrational modes.