High-throughput directed evolution of affinity proteins
Development of new affinity proteins using combinatorial protein engineering is today established for generation of monoclonal antibodies and also essential for discovery of binders that are based on non-immunoglobulin proteins. Phage display is most frequently used, but yeast display is becoming increasingly popular, partly due to the option of utilizing fluorescence-activated cell sorting (FACS) for isolation of new candidates. Escherichia coli has several valuable properties for library applications and in particular the high transformation efficiency.
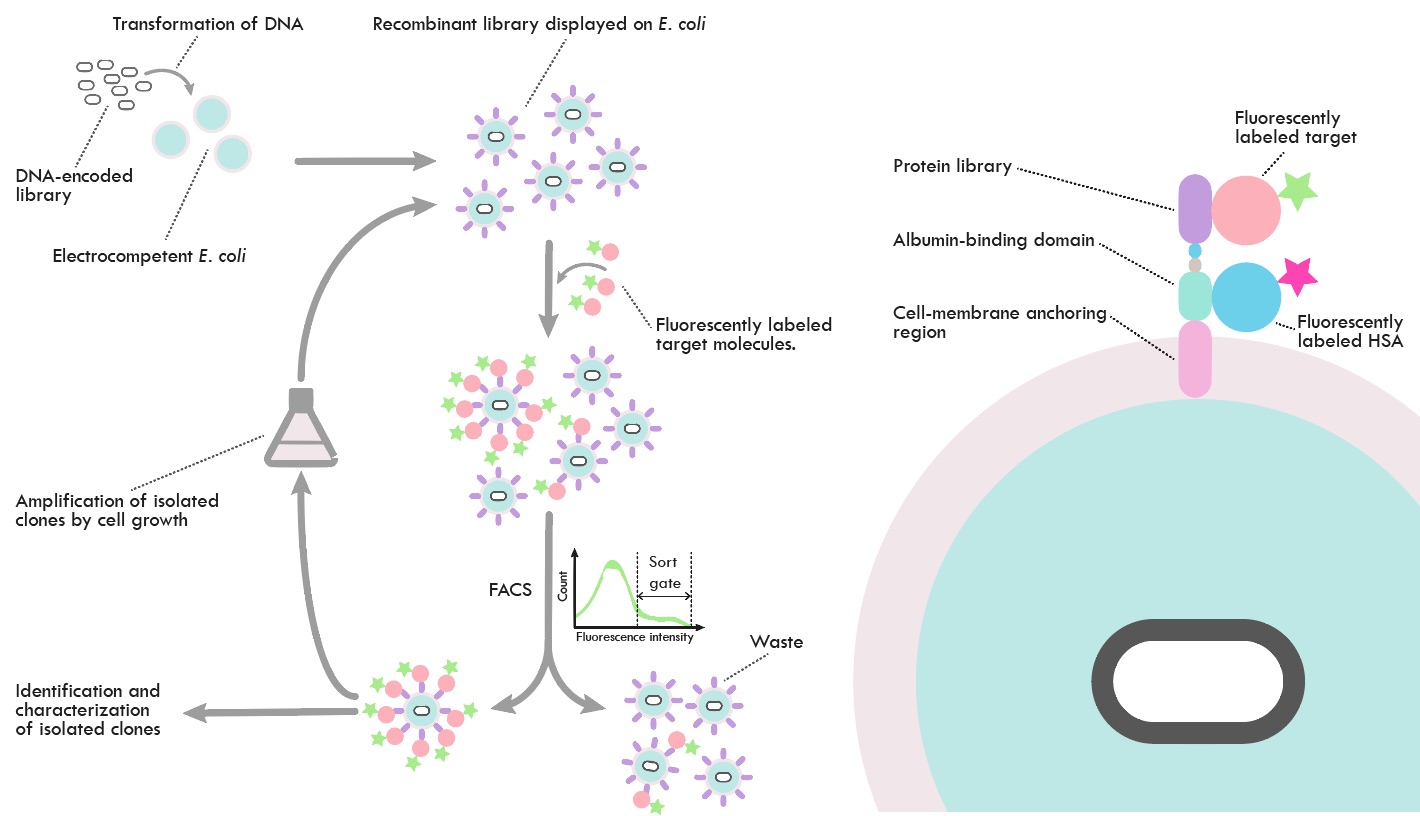
The project is focused on development of a new E. coli-based bacterial surface display system for proteins and its use in selection of protein-based affinity reagents from combinatorial libraries. In the project, display of proteins on the E. coli surface is based on so called autotransporters, naturally occurring proteins characterized by a capability to direct and present "passenger" proteins to the outside of the cell via an anchoring domain positioned in the outer membrane. Preliminary data support the use of this system for protein library applications, and a number of potential improvements have been identified, which are addressed in the present project to develop the AIDA-I surface display system as a viable option for protein display application of relevance. These issues include throughput, cell-cell aggregation and optimizations to allow surface display of a wide range of affinity proteins classes of different structure and molecular properties. The project is part of the CellNova VINNOVA center.
Research funded by: Vinnova - Sweden's innovation agency