Regulatory Genomics
Our group's research aims to discover and analyse the functional part of the non-coding genome to enable its clinical translation and understand its role in evolution.
Research
Around 35 percent of our genome are unique sequences but do not code for any protein or not strongly conserved. The mission is to understand the contribution of the non-coding genome towards organism well-being, survival and health.
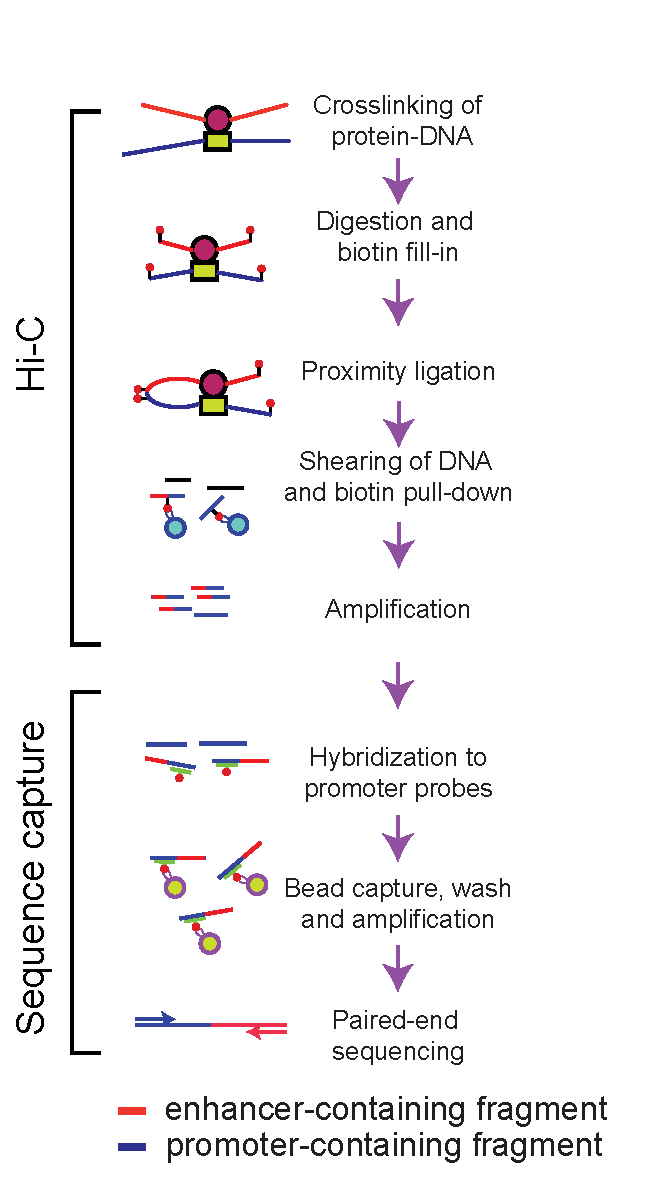
We use chromatin looping information to map promoter-enhancer interactions in various tissues, cells and organisms. We developed a method that provides high-resolution promoter-enhancer maps (Capture Hi-C, HiCap, Chi-C) (Sahlén P. et al, 2015). We also use chromatin immunoprecipitation, gene expression data to refine interaction maps, and develop experimental and computational methods for functional characterisation of enhancers.
Genome-wide association studies (GWAS) revealed that almost all variants associated with complex diseases such as cardiovascular disease is non-coding. Although two-thirds of such variants are within regulatory regions, it is not a straightforward task to determine the gene(s) they regulate. This, in turn, hampers the clinical translation of genome information for the management of such diseases. In our lab, we develop and apply 3D genome and epigenome information to understand the genetic component of complex diseases, currently focusing on atherosclerosis, aneurysm and inflammatory skin diseases (Åkerborg Ö., 2019).
The domestication of animals and plants was the basis of the transition from hunting and gathering to farming. The domestic dog is considered the first domesticated animal and is unique in its integration in the human society. We use chromatin interaction and epigenomics to discover the role of non-coding variation in the evolution of domestic dog from wolves.
