An Algorithm to Track Cell Movements
News
PhD student Klas Magnusson has developed a successful new algorithm for automated image analysis in cell biology. Tests using the algorithm at Stanford University show that it can reduce manual labor by more than 90 per cent. Magnusson was recently awarded a “Best Student Paper” award for his creation.
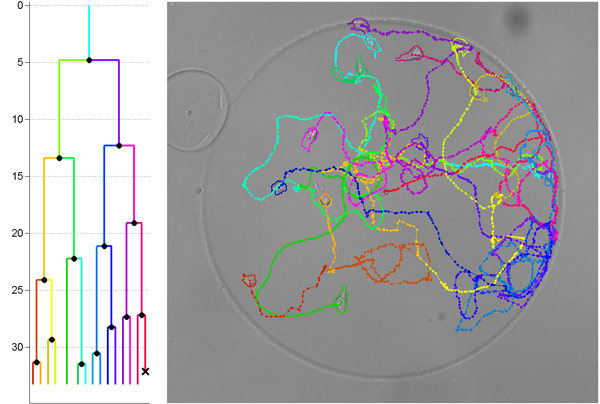
Cell biologists often acquire long sequences of microscope images, which must then be processed image-by-image to annotate numbers of cells, shapes, velocities and other parameters. It’s a tedious and time-consuming procedure.
An automated solution could speed up research across entire field of cell biology by cutting down on manual labor, making the work faster, more scalable and more quantitative. That’s precisely what Klas Magnusson is doing by algorithm.
“We can reduce manual labor by over 90 per cent when tracking cells in image sequences, depending on what parameters we’re looking for,” he says. “This gives us reliable data analysis and expands our capacity for biomedical experimentation. We believe we can ultimately cut out manual handling altogether.”
Magnusson has co-written a paper presenting a new algorithm that links cell detections in an image sequence into “tracks” — or sequences of positions that define where a cell has been at different points in time. Similar algorithms were originally developed for applications such as real-time tracking of aircraft on radar screens, but applied to cell data analysis, these algorithms process sequences one image at a time, which is far from ideal. In cell tracking applications, the entire image sequence is usually available when the processing starts. Magnusson’s algorithm takes advantage of this by repeatedly adding new cell tracks one at a time.
“Our new algorithm is different from most existing ones in that it simultaneously looks at every image in the sequence. This allows it to make much more reliable decisions regarding cell movement,” Magnusson explains.

The algorithm has already been used to identify better culturing conditions for stem cells, which many researchers expect to soon contribute to treatments for muscular dystrophy, leukemia and other serious diseases. Magnusson believes algorithms of this type will eventually become as important for cell biologists as microscopes and DNA sequencing are today. “The goal is to release the algorithms that we develop in open source software, so that biologists all over the world can use them,” he says.
But Magnusson’s algorithm isn’t limited to tracking cells. It might eventually be applied to problems beyond cell biology, such as revealing the movements of people, cars, clouds, or other objects in images.
The cell tracking project in the Signal Processing Lab at KTH’s School of Electrical Engineering is carried out in close collaboration with the Blau Lab at Stanford University in the U.S., where algorithms developed at KTH are used in biological research.
Klas Magnusson received a Best Student Paper Award at the 2012 International Symposium on Biomedical Imaging in Barcelona, Spain for “A batch algorithm using iterative application of the Viterbi algorithm to track cells and construct cell lineages.” Co-written with KTH Assistant Professor Joakim Jaldén, it’s the first publication originating in a newly started project in the Signal Processing Lab that aims to develop automated image analysis algorithms for applications in cell biology. Financing for the project is provided by the Swedish Research Council (Vetenskapsrådet).
By Marie Androv. Edited by Kevin Billinghurst.

